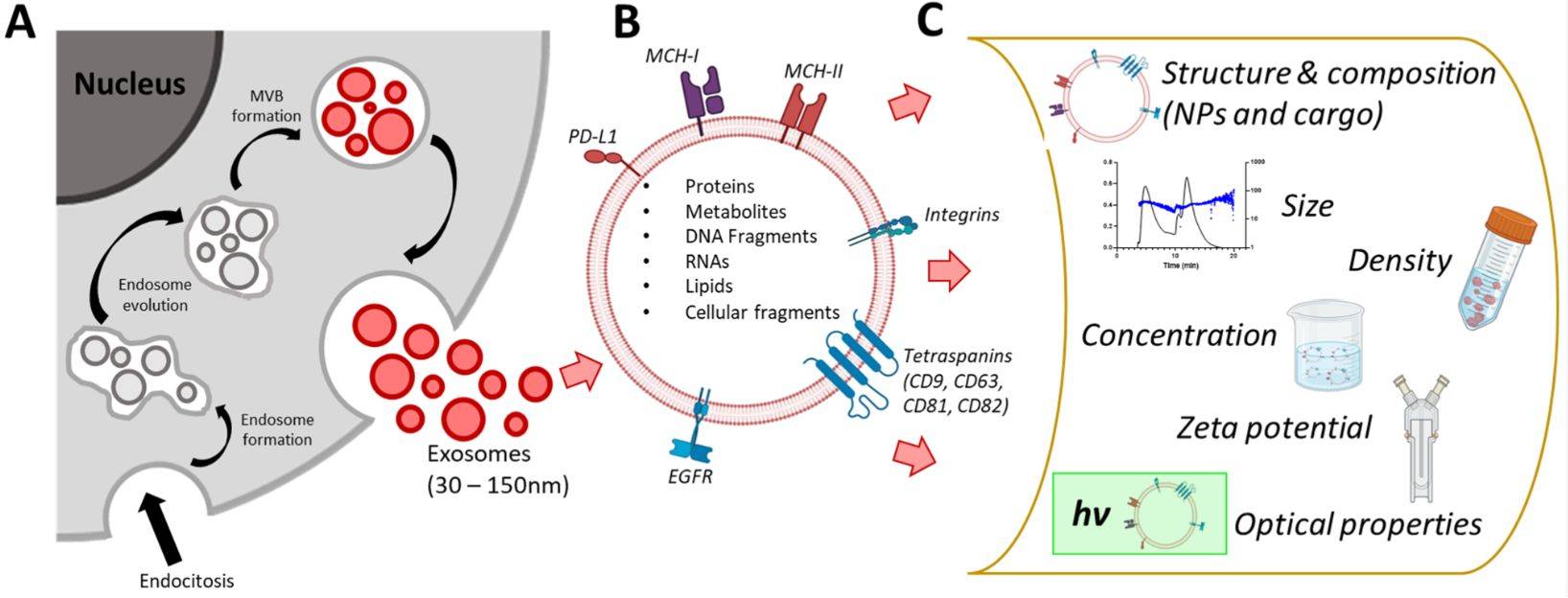
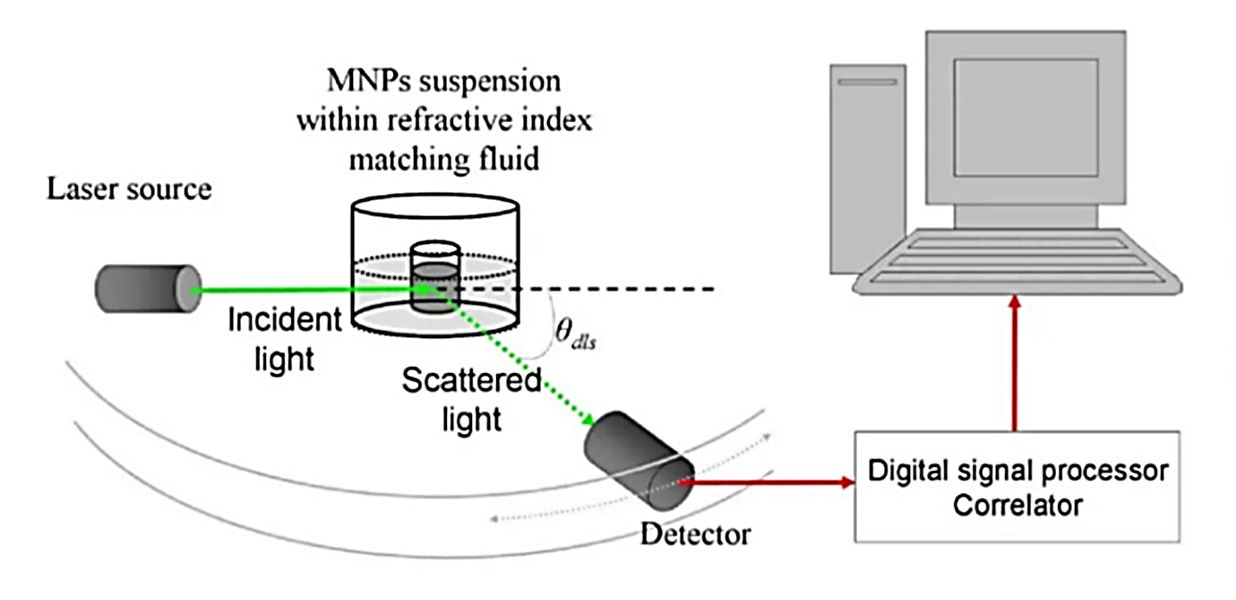
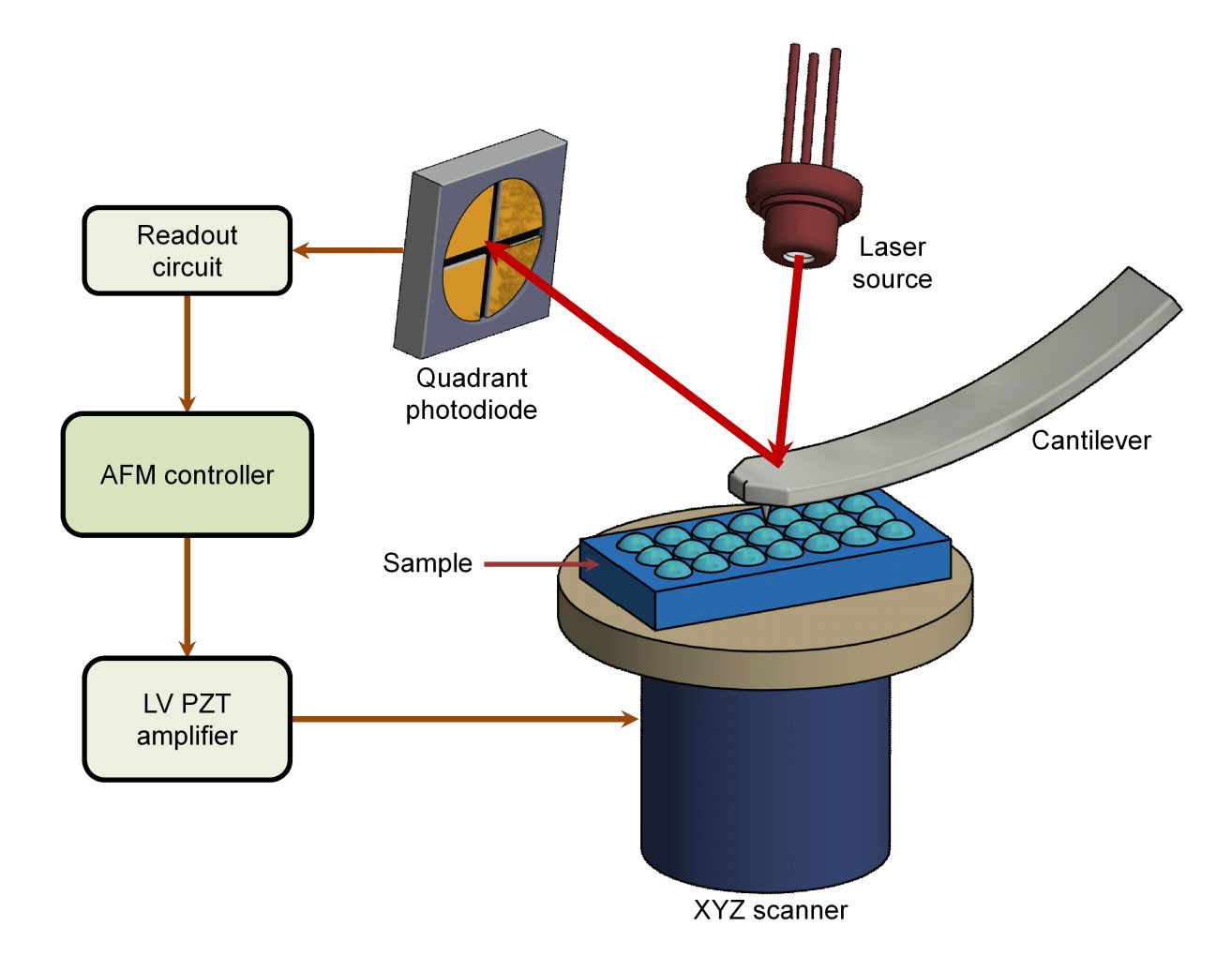
Exosome Surface Marker Analysis Service
Exosome surface marker analysis is essential for confirming the identity, purity, and biological relevance of extracellular vesicle (EV) preparations. Exosomes are nanosized vesicles, typically 30-150 nm in diameter, secreted by nearly all cell types and present in a wide range of biological fluids. They encapsulate proteins, lipids, and nucleic acids that reflect the molecular profile of their cell of origin.
Surface proteins such as CD9, CD63, CD81, TSG101, and ALIX are widely recognized as hallmark exosome markers, providing critical information for both basic research and translational applications. At Creative Biostructure, our high-quality, customizable exosome surface marker analysis services follow the latest Minimal Information for Studies of Extracellular Vesicles (MISEV2023) guidelines to ensure scientific rigor and reproducibility.
Why Is Protein Composition Essential in Exosome Characterization?
The protein composition of exosomes is a key indicator of their origin, quality, and functional potential. According to the MISEV2023 framework, protein-based EV characterization involves assessing:
- Transmembrane or GPI-anchored proteins linked to the plasma membrane or endosomes (e.g., tetraspanins CD9, CD63, CD81).
- Cytosolic proteins that are often enriched in EVs (e.g., TSG101, ALIX).
- Negative or purity control markers from non-EV contaminants (e.g., albumin, apolipoproteins).
- Intracellular compartment markers (e.g., histones, mitochondrial proteins) when relevant to the study.
- Secreted proteins that may associate with the EV surface ("corona" proteins).
Comprehensive analysis of these exosome surface proteins is critical to:
- Validate vesicle identity
- Confirm sample purity
- Support biomarker discovery
- Guide therapeutic development
- Ensure manufacturing quality control in research and industrial applications
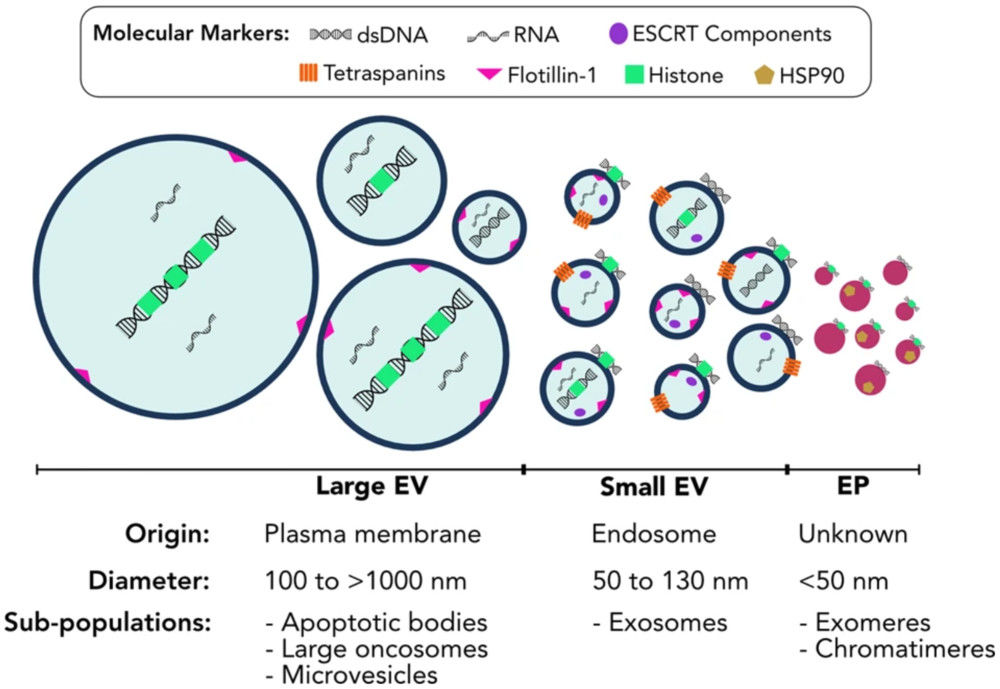
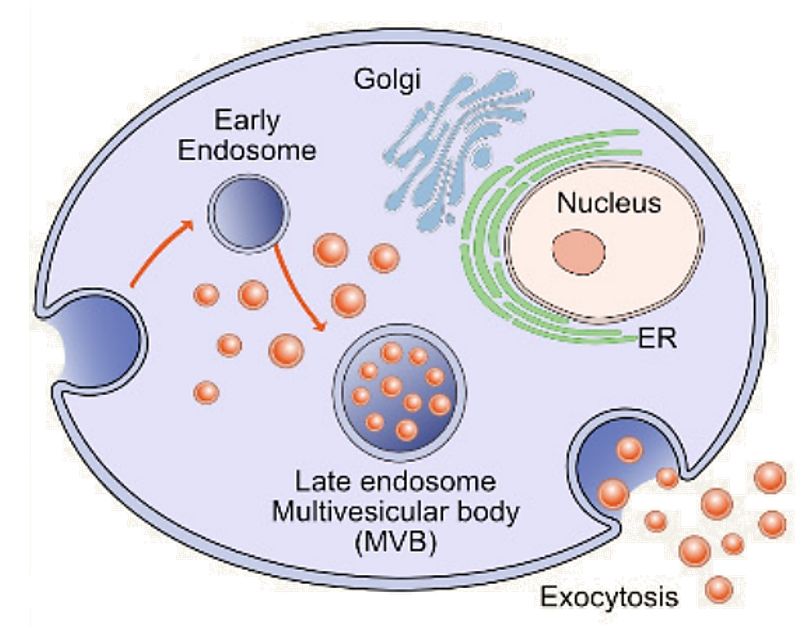
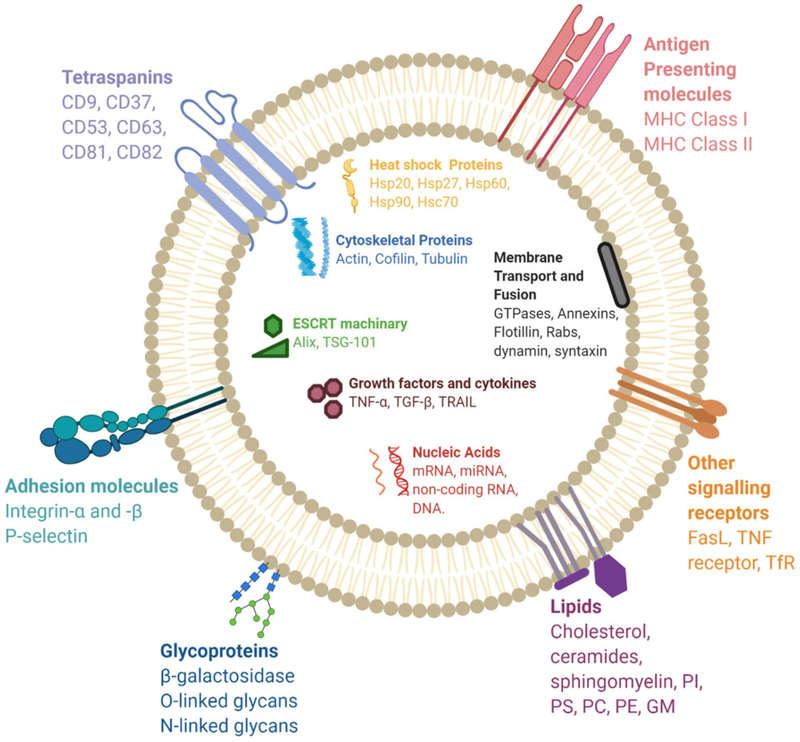 Figure 1. Molecular Composition of Exosomes. (Gurung S, et al., 2021)
Figure 1. Molecular Composition of Exosomes. (Gurung S, et al., 2021)
Techniques We Offer for Exosome Surface Marker Profiling
Creative Biostructure's exosome surface marker profiling integrates multiple analytical platforms to provide accurate, reproducible, and publication-ready results. Our capabilities include:
| Method | Detection Principle | Key Strengths | Limitations | Best For | Turnaround Time |
|---|---|---|---|---|---|
| Western Blotting | Separation of exosomal proteins via SDS-PAGE, followed by antibody-based detection of specific exosome surface proteins | - Highly specific marker detection (CD9, CD63, CD81, TSG101, ALIX) - Established, widely accepted method - Qualitative confirmation of vesicle identity |
- Low throughput - Semi-quantitative only |
Confirming exosome specific markers according to MISEV2023 | 5-7 business days |
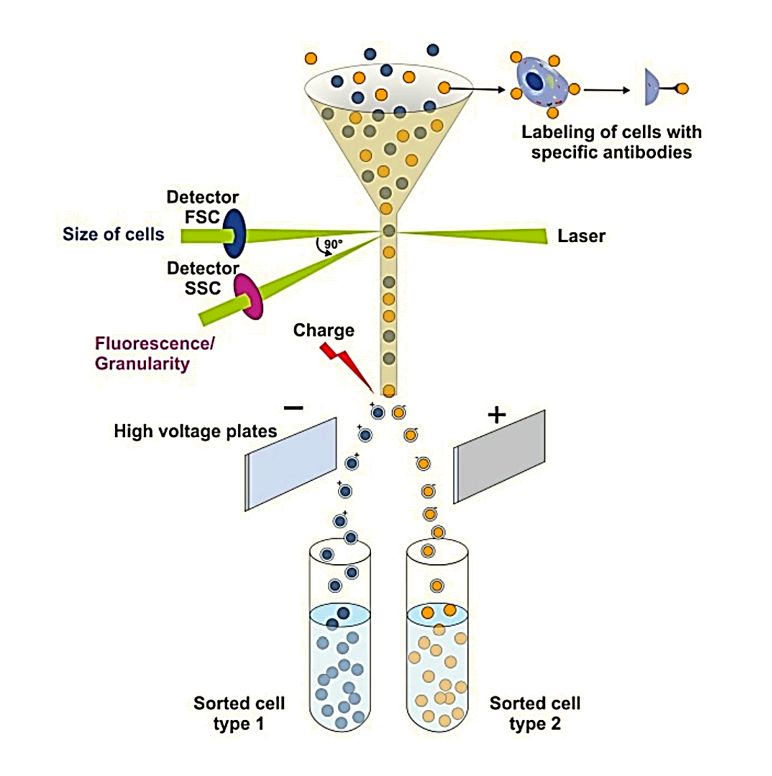
| Nano-Flow Cytometry | Single-particle analysis using fluorescence-labeled antibodies targeting exosome markers | - Quantitative profiling of individual EVs - Can analyze subpopulations - High throughput |
- Requires high-quality antibodies - May have size detection limits | Rapid exosome surface markers profiling and subpopulation analysis | 3-5 business days |
| ELISA | Antibody-based capture and detection of target exosome surface proteins in bulk EV samples | - High sensitivity and specificity - Suitable for absolute quantification - Scalable for multiple samples |
- Limited to known target proteins - Cannot assess particle heterogeneity |
Quantitative analysis of selected exosomes markers in research and QC | 3-5 business days |
| Mass Spectrometry (Proteomics) | High-resolution protein identification and quantification via LC-MS/MS | - Comprehensive marker discovery - Unbiased identification of known & novel proteins - High sensitivity |
- Requires specialized sample prep - Longer turnaround time |
Biomarker discovery, in-depth exosome surface proteins mapping | 2-6 weeks |
We offer custom marker detection, enabling analysis of client-specified exosome specific markers beyond the commonly used panel.
Our Workflow for Exosome Surface Marker Analysis
Consultation & Project Planning
Define objectives, select target exosome markers, and choose analytical methods.
Sample Submission
Provide prepared EV samples or source material for isolation.
Laboratory Analysis
Conduct marker detection using agreed platforms.
Data Processing
Compile and interpret results in accordance with MISEV2023.
Report Delivery
Provide a comprehensive analytical report with raw data and visual summaries.
 Figure 2. Project Workflow for Exosome Surface Marker Detection. (Creative Biostructure)
Figure 2. Project Workflow for Exosome Surface Marker Detection. (Creative Biostructure)
Sample Requirements and Compatibility
| Sample Type | Minimum Amount | Preparation & Storage | Shipping Conditions |
|---|---|---|---|
| Purified Exosomes | ≥ 100 µL | Store in PBS or suitable buffer at -80 °C; avoid repeated freeze-thaw cycles | Ship on dry ice |
| Plasma / Serum / Urine | ≥ 1 mL | Freeze immediately at -80 °C after collection | Ship on dry ice |
| Cell Culture Supernatant | ≥ 5 mL | Remove cells and debris via centrifugation; store at -80 °C | Ship on dry ice |
| Other Biological Fluids* | Contact us | Follow matrix-specific preparation guidelines | Ship on dry ice |
* Examples include saliva, cerebrospinal fluid (CSF), bile, breast milk, amniotic fluid, plant-derived fluids, and tissue homogenates. For uncommon sample types, please contact us for tailored instructions.
What Deliverables Will You Receive
- Qualitative and/or quantitative results for selected exosome surface markers.
- Detailed methodology and experimental conditions.
- Data interpretation and visual representation (graphs, blots, flow plots).
- Raw data files for in-depth analysis.
How Can Exosome Surface Marker Analysis Be Applied
Our exosome surface marker analysis services are widely used in:
- Disease biomarker research: cancer, neurodegenerative, cardiovascular, and infectious diseases.
- Exosome-based drug development and therapeutic monitoring.
- Quality control for exosome isolation and manufacturing workflows.
- Fundamental research in cell biology and intercellular communication.
Why Choose Creative Biostructure
- Compliance with MISEV2023 for scientifically robust reporting.
- Expert team with years of EV characterization experience.
- Advanced instrumentation for precise and reproducible results.
- Flexible and customizable services for both research and industrial needs.
- Proven track record with global academic and biotech clients.
Case Study
Case: Characterization of hUC-MSC-Derived Exosomes for Targeted Anti-miR-146b Delivery in Colorectal Cancer
Background
This study explored human umbilical cord mesenchymal stem cell (hUC-MSC)-derived exosomes as delivery vehicles for a phosphorodiamidate morpholino oligonucleotide (PMO) targeting miR-146b-5p in colorectal cancer (CRC). To ensure specificity, PMO was anchored to exosomal membranes via the CD63-binding CP05 peptide, making exosome surface marker analysis critical for validating vesicle identity and targeting capability.
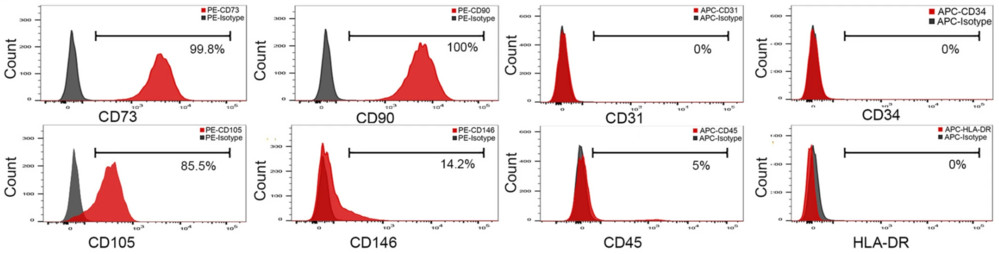 Figure 3. Flow cytometry analysis evaluating the expression of key surface markers, including CD73, CD90, CD105, CD146, CD31, CD34, and CD45. (Yu S, et al., 2024)
Figure 3. Flow cytometry analysis evaluating the expression of key surface markers, including CD73, CD90, CD105, CD146, CD31, CD34, and CD45. (Yu S, et al., 2024)
Methods
- Isolation & Verification: Exosomes were purified by ultracentrifugation and characterized using TEM, nanoflow cytometry, and Western blot.
- TEM confirmed typical vesicle morphology (~69 nm).
- Western blot detected exosomal markers CD63, CD81, TSG101 and absence of contamination markers Syntaxin6 and Erp72.
- Flow cytometry showed high CD63 (86.9%), CD73 (71.9%), and CD90 (88.2%) expression, confirming MSC origin.
- Surface Marker Binding Optimization: Flow cytometry verified efficient CP05-PMO binding to CD63-positive
Results
- CP05-PMO conjugation preserved exosomal surface marker profiles while enabling efficient cargo loading.
- In vitro, ePPMO-146b reduced CRC cell viability (61.28% in SW620, 71.00% in Caco2) and inhibited migration by up to 90.05%.
- Marker analysis revealed EMT suppression, with increased E-cadherin and decreased N-cadherin/vimentin.
- In vivo, ePPMO-146b accumulated in tumors, reduced volume by 71.12%, and showed no systemic toxicity.
Conclusion
Rigorous exosome surface marker analysis confirmed the purity, identity, and MSC origin of engineered vesicles, ensuring targeted delivery efficiency and safety. This highlights the essential role of surface marker profiling in developing exosome-based therapeutics for cancer treatment.
At Creative Biostructure, we combine cutting-edge technology with expert scientific insight to deliver precise, reproducible exosome surface marker analysis results. Whether you are validating vesicle identity, discovering novel biomarkers, or ensuring manufacturing quality, our team provides tailored solutions that align with the latest MISEV2023 standards. Contact us to discuss your project and receive a customized proposal.
References
- Gurung S, Perocheau D, Touramanidou L, et al. The exosome journey: from biogenesis to uptake and intracellular signalling. Cell Communication and Signaling. 2021, 19(1): 47.
- Welsh J A, Goberdhan D C I, O'Driscoll L, et al. Minimal information for studies of extracellular vesicles (MISEV2023): From basic to advanced approaches. Journal of Extracellular Vesicles. 2024, 13(2): e12404.
- Yu S, Liao R, Bai L, et al. Anticancer effect of hUC-MSC-derived exosome-mediated delivery of PMO-miR-146b-5p in colorectal cancer. Drug Delivery and Translational Research. 2024, 14(5): 1352-1369.
Frequently Asked Questions
For any inquiries, our support team is ready to help you get technical support for your research and maximize your experience with Creative Biostructure.