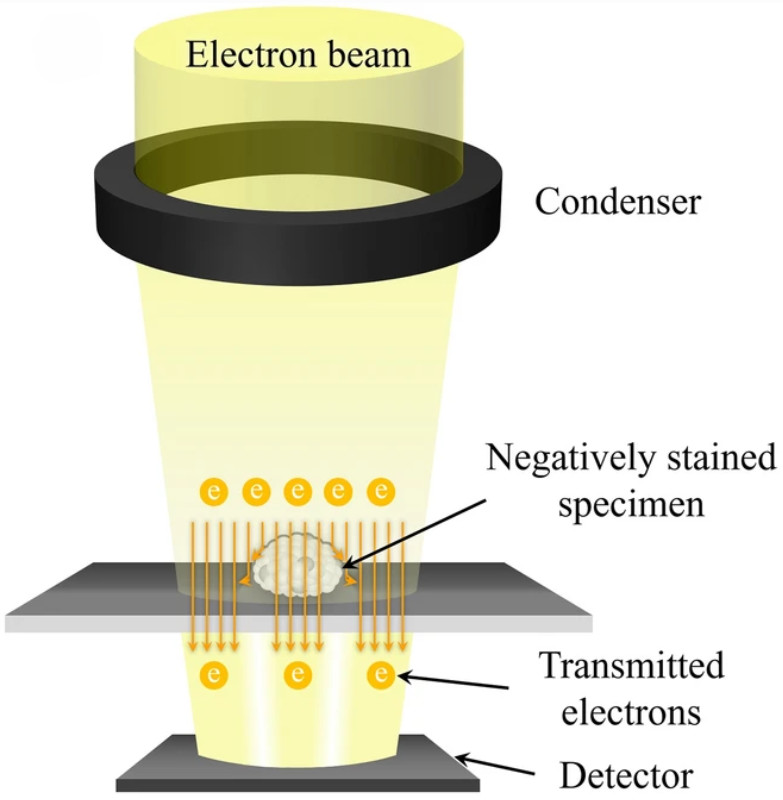
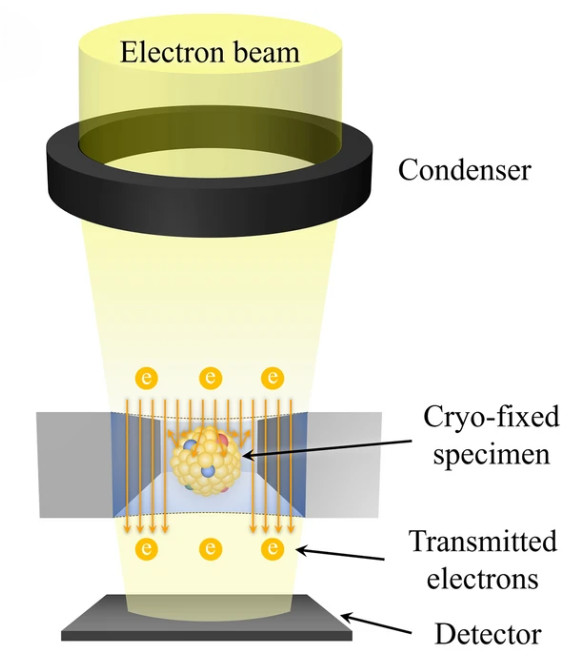
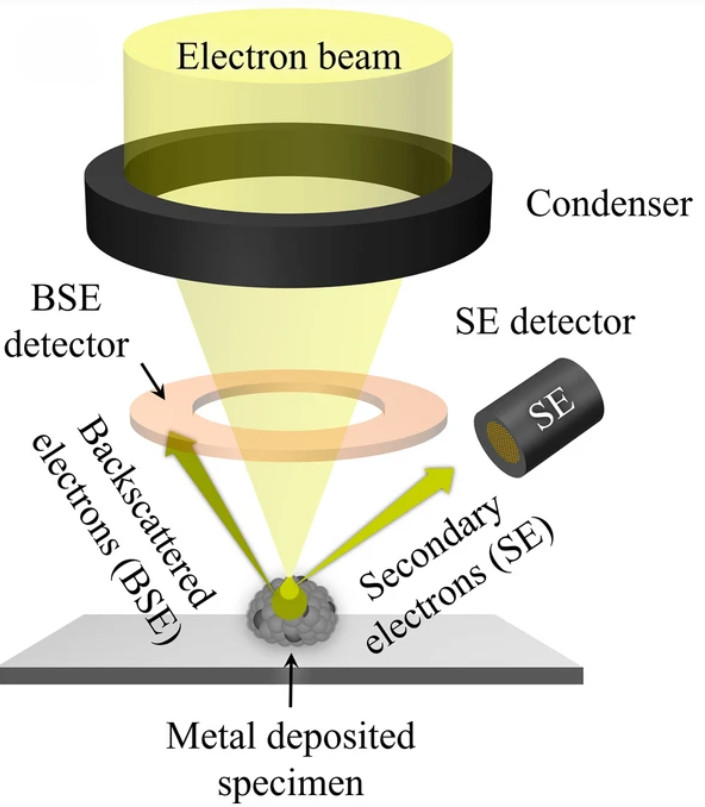
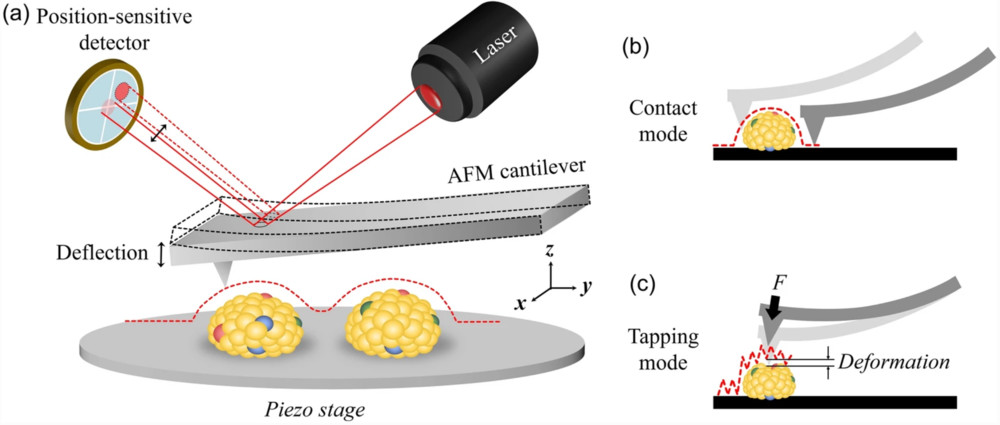
Mass Spectrometry-Based Exosome Characterization Service
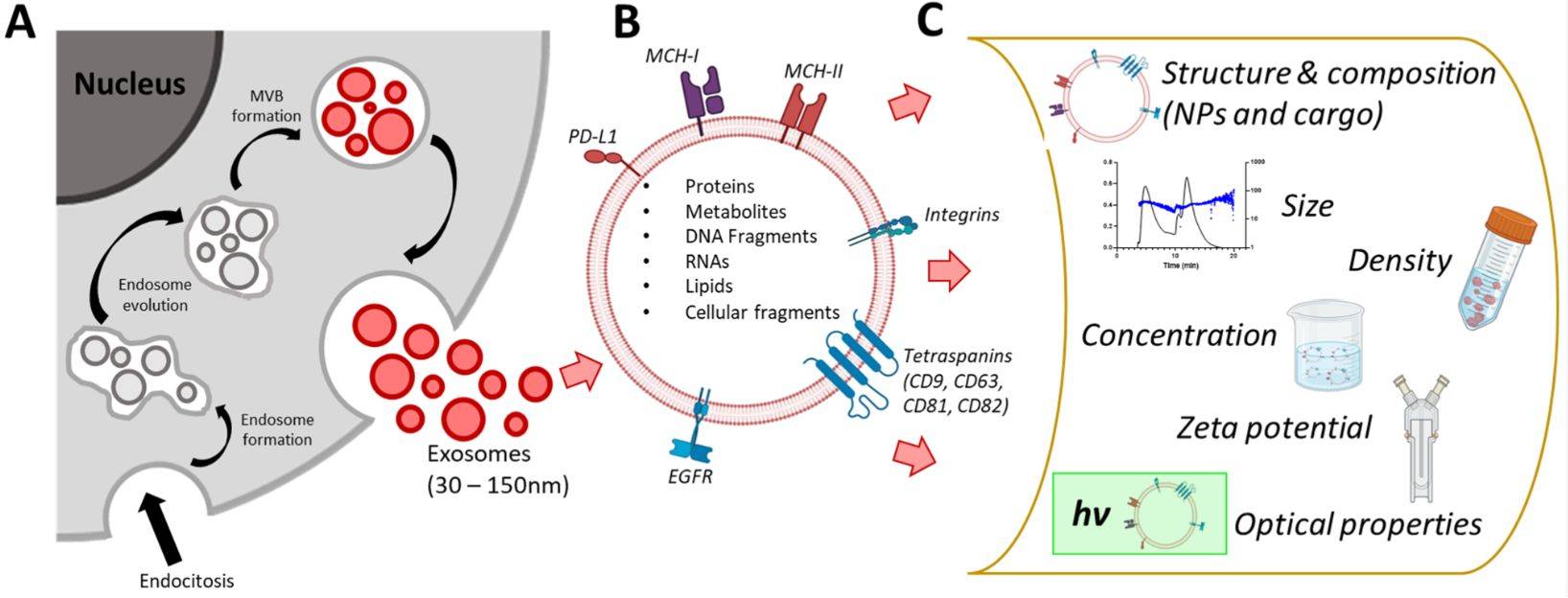
Extracellular vesicles (EVs), including exosomes, are pivotal mediators of intercellular communication, carrying complex molecular cargo that reflects the physiological state of their parent cells. To unlock their immense potential in diagnostics and therapeutics, it is essential to deeply characterize this cargo.
High-resolution liquid chromatography-tandem mass spectrometry (LC-MS/MS) stands as the gold-standard technology for this task. It offers the sensitivity, specificity, and scale required for a comprehensive and unbiased mass spectrometry analysis of exosomes. At Creative Biostructure, we provide an end-to-end service that transforms your EV samples into actionable biological insights, empowering the next wave of scientific discovery.
Why Use Mass Spectrometry for Exosome Characterization?
Mass spectrometry (MS) is one of the most powerful technologies for analyzing the exosome proteome and other molecular components. By measuring mass-to-charge ratios with high precision, MS enables:
- Comprehensive Profiling: Identification of proteins, lipids, and metabolites, including low-abundance species.
- Quantitative Analysis: Accurate measurement of relative and absolute abundances.
- Targeted and Untargeted Approaches: Discovery-based proteomics for biomarker exploration, and targeted LC-MS/MS for quantification of defined proteins.
- Multi-omics Integration: Proteomics, lipidomics, and metabolomics datasets can be combined to provide a holistic view of exosomal function.
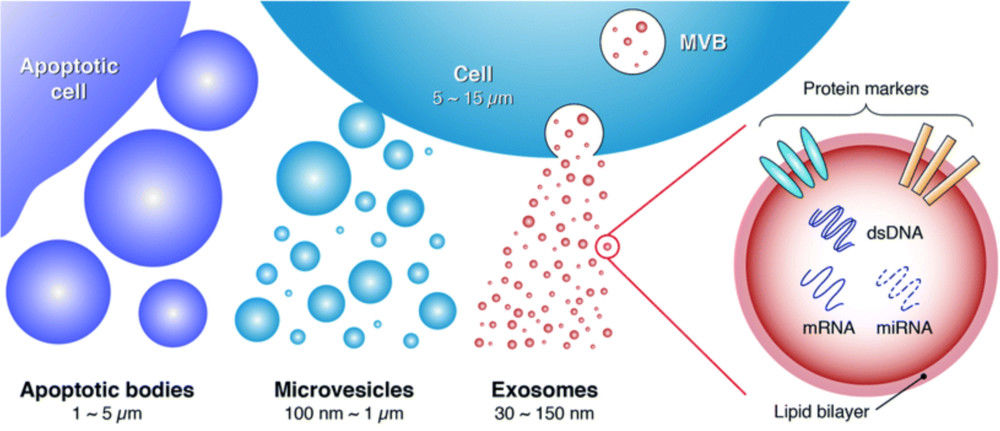
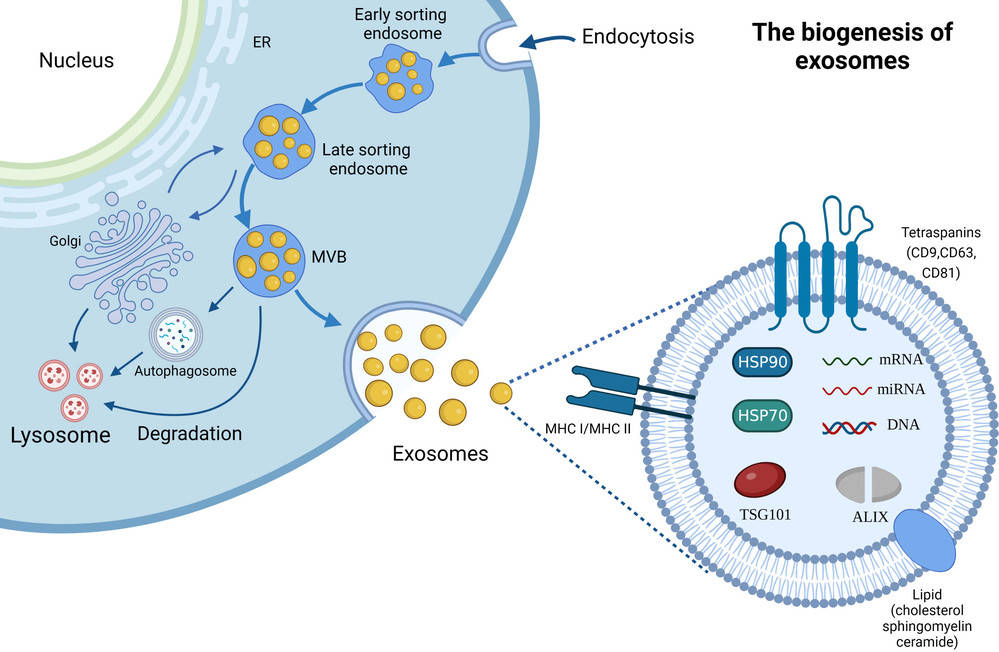
Frequently studied exosomal proteins include CD9, CD63, CD81, ALIX, and TSG101, which serve as vesicle identity markers, as well as disease-associated proteins relevant to oncology, neurology, and immunology.
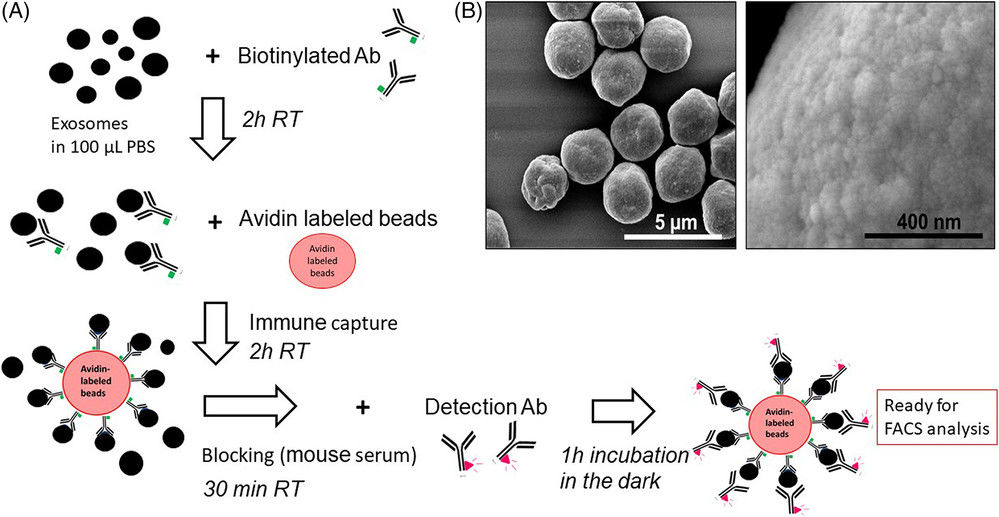
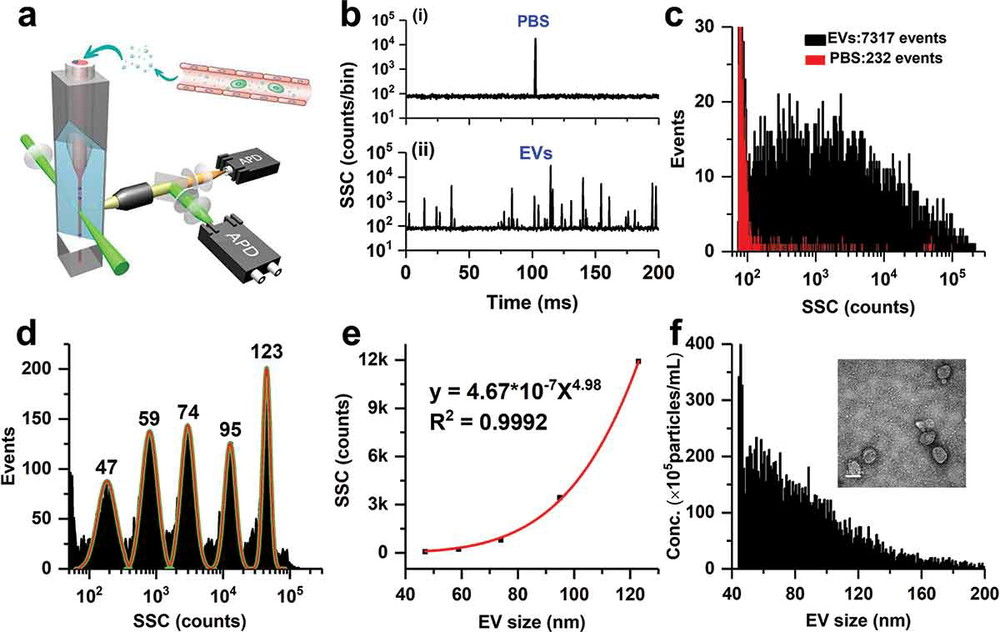
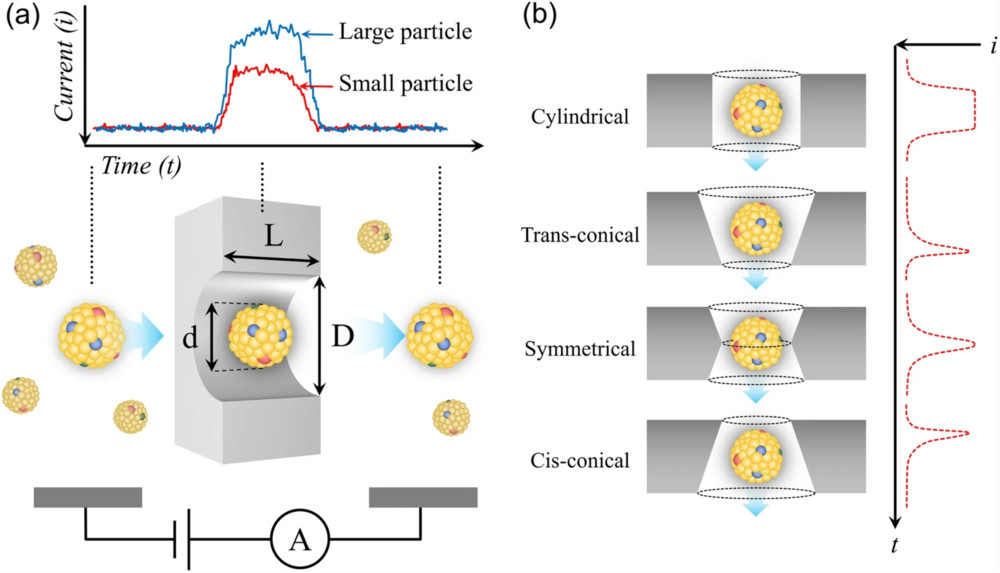
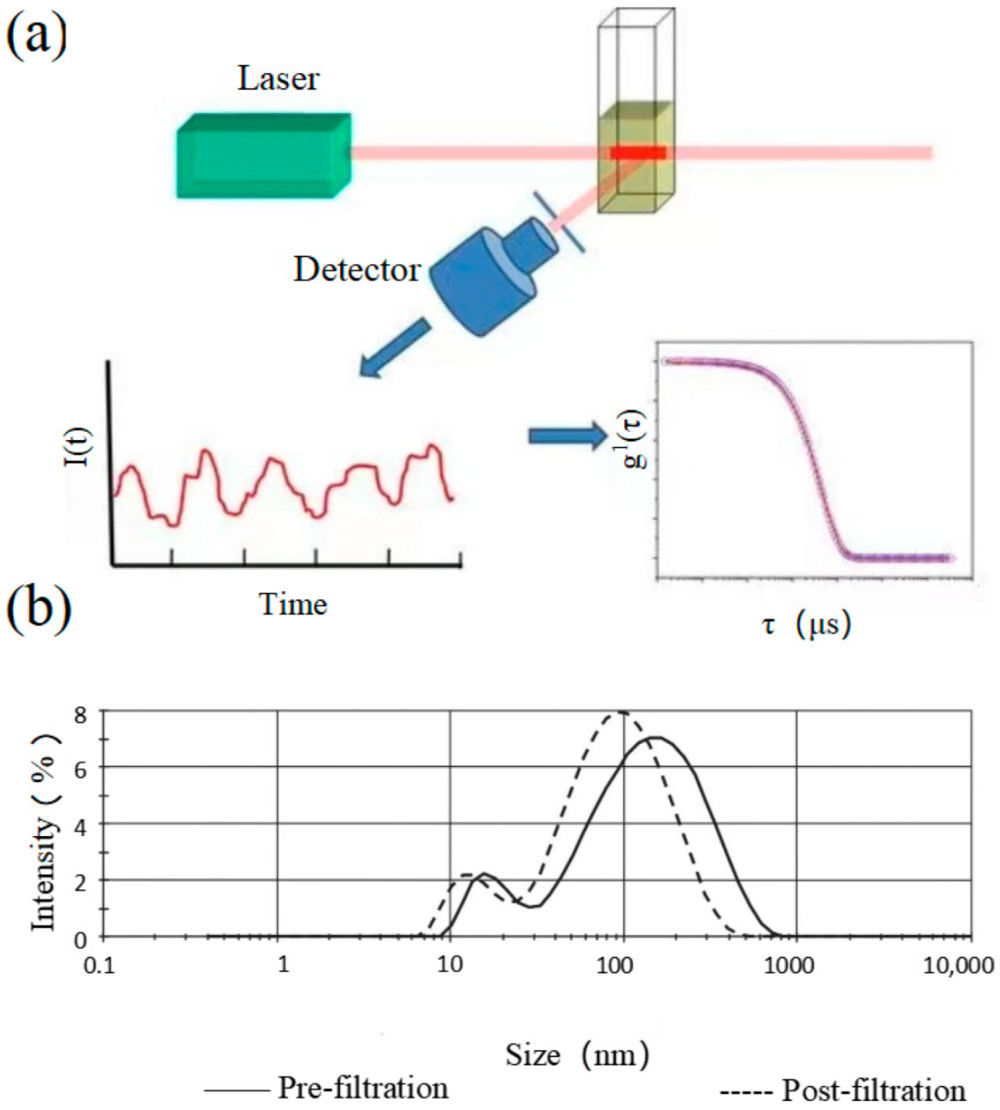
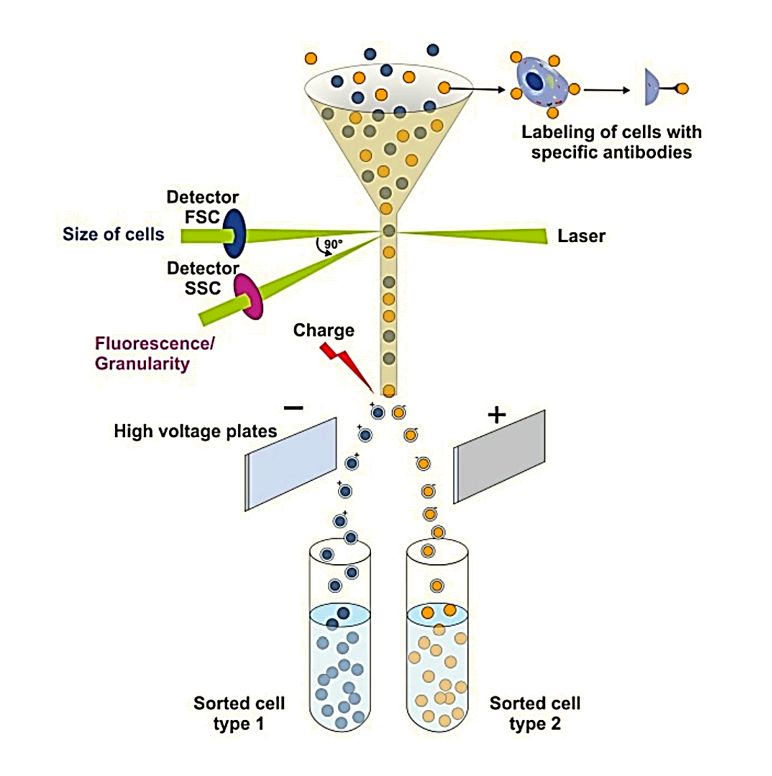
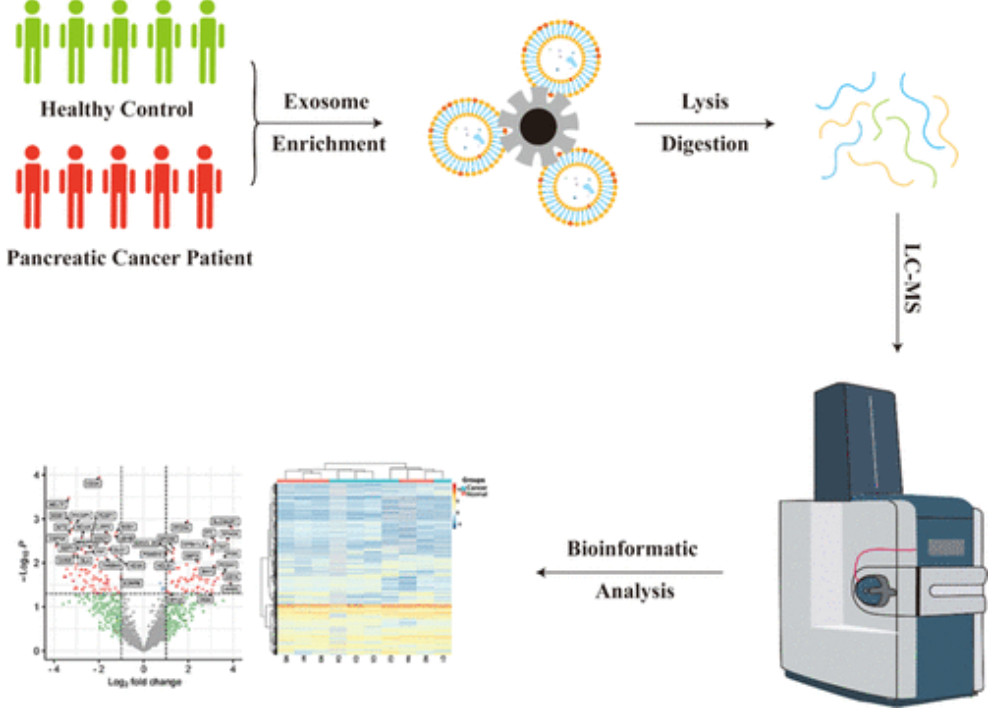 Figure 1. Schematic of urinary exosome detection in cancer patients using mass spectrometry. (Cheng X, et al., 2024)
Figure 1. Schematic of urinary exosome detection in cancer patients using mass spectrometry. (Cheng X, et al., 2024)
Our Comprehensive Multi-Omics Services
We offer a suite of specialized mass spectrometry services to provide a holistic view of your exosome and EV samples.
Exosome Proteomics
Characterize the complete protein landscape of your EVs to identify biomarkers and understand functional pathways.
- Untargeted (Discovery) Proteomics: Ideal for global protein profiling and novel biomarker discovery. Utilizing state-of-the-art Orbitrap LC-MS/MS platforms, we perform deep proteomic screening to identify thousands of proteins, providing a comprehensive map of the exosome proteome.
- Targeted Proteomics: Designed for the absolute quantification and validation of specific proteins of interest. Using highly sensitive Triple Quadrupole (QQQ) MS with Multiple Reaction Monitoring (MRM), this service is perfect for validating biomarkers and analyzing post-translational modifications.
Exosome Lipidomics
Analyze the diverse lipid composition of EV membranes and cargo. Lipid profiles are critical for understanding EV biogenesis, stability, cell targeting, and signaling functions.
Exosome Metabolomics
Identify and quantify the small-molecule metabolites within exosomes. This analysis offers a snapshot of the metabolic state of the originating cells and can reveal novel pathways in disease.
Mass Spectrometry Platforms for Exosome Analysis
| Platform | Key Advantages |
|---|---|
| Orbitrap MS | High-resolution detection for untargeted exosome proteomics |
| Triple Quadrupole LC-MS/MS | Targeted, quantitative assays for peptides and proteins |
| Q-TOF MS | Fast, sensitive analysis of exosomal proteins and metabolites |
| MALDI-MS | High-throughput screening of exosome-associated biomolecules |
All instruments are maintained with constant parameters to ensure reproducibility. Each project includes positive and negative controls, and SIL peptide standards are spiked into samples for validation.
Mass Spectrometry Exosome Characterization Workflow
Project Consultation & Strategic Design
Our process begins with a detailed consultation. We collaborate with you to understand your specific research objectives, designing a tailored analytical strategy that is optimized to deliver the most meaningful and impactful results.
Sample Integrity Check & EV Isolation
Upon receiving your materials, we conduct rigorous initial quality control. We then employ state-of-the-art isolation techniques, ensuring that your resulting extracellular vesicle population is of the highest purity and integrity for sensitive downstream analysis.
Multi-Omic Sample Preparation
Our specialized protocols prepare your pure EV samples for proteomic, lipidomic, or metabolomic interrogation. This crucial phase involves meticulous extraction and enzymatic digestion to ready the molecular components for detection by the mass spectrometer.
Advanced Mass Spectrometry Acquisition
Your samples are analyzed using our cutting-edge LC-MS/MS platforms. We generate high-resolution, high-mass-accuracy data, capturing a deep and precise profile of the molecular species and their relative abundances within your samples.
In-Depth Bioinformatic Analysis
Raw data is transformed into biological insight by our expert bioinformatics team. We perform comprehensive statistical analysis, functional annotation, and pathway enrichment to uncover the significant patterns and stories hidden within your dataset.
Comprehensive Reporting & Expert Consultation
The project concludes with the delivery of a publication-ready report detailing all methods, data, and findings. We then schedule a final consultation to review the results with you, ensuring you can fully interpret and leverage the data for your research.
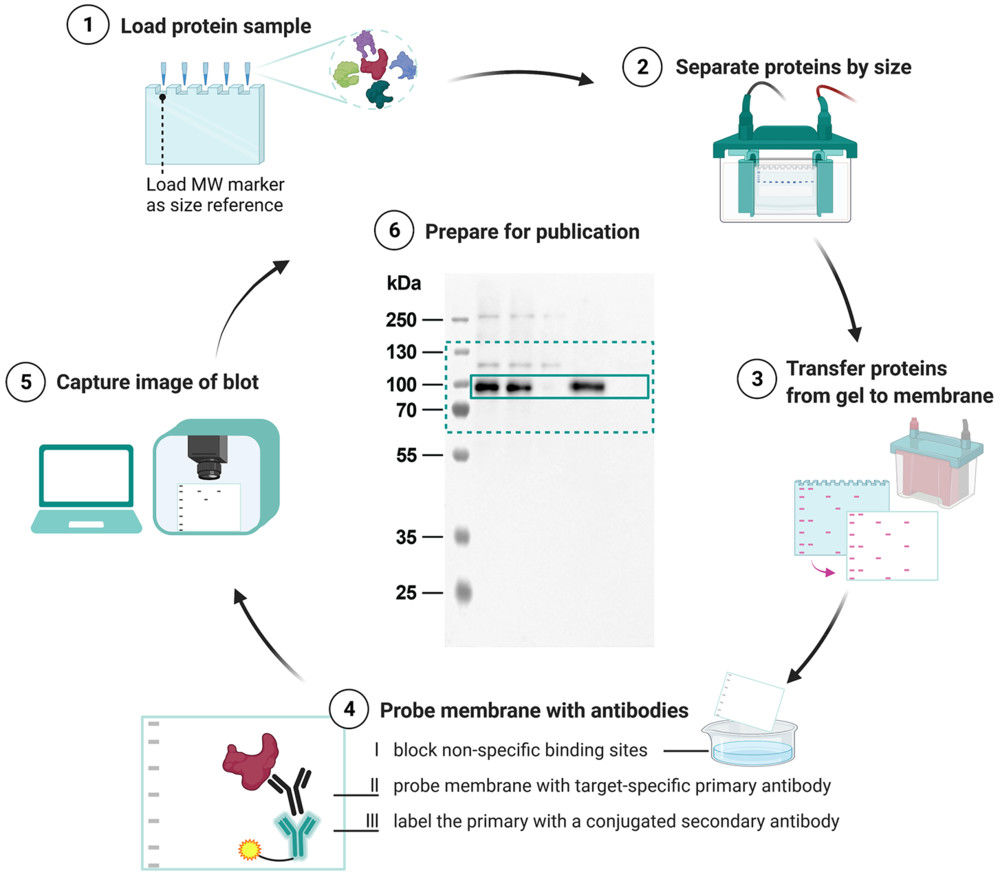
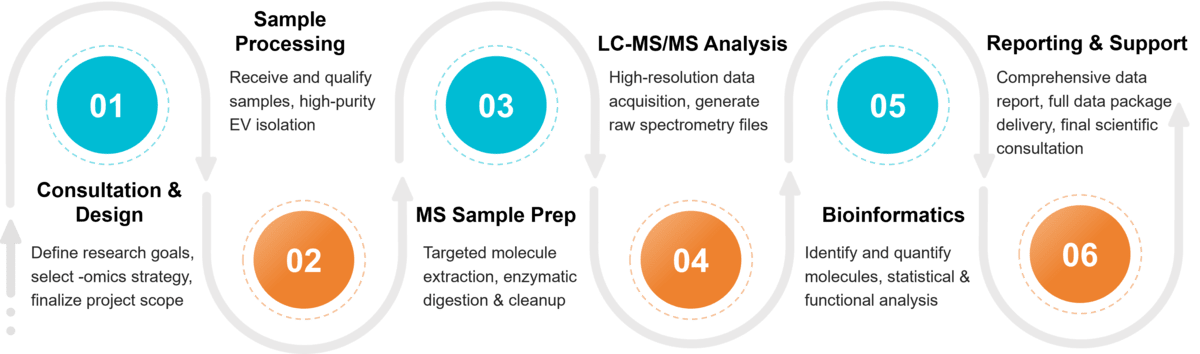 Figure 2. Project Workflow for Exosome Characterization by Mass Spectrometry. (Creative Biostructure)
Figure 2. Project Workflow for Exosome Characterization by Mass Spectrometry. (Creative Biostructure)
Sample Requirements for Mass Spectrometry
To ensure high-quality and reproducible results, please follow the recommended sample submission guidelines below:
| Sample Type | Recommended Volume / Amount | Notes |
|---|---|---|
| Pre-purified exosomes | ≥ 50-100 µg total protein (or ≥ 1×109 particles) | Provide in low-salt, MS-compatible buffer; avoid detergents |
| Plasma / Serum | 0.5-2 mL | Collect in EDTA or heparin tubes; avoid repeated freeze-thaw cycles |
| Urine | 5-10 mL | Fresh or frozen samples; protease inhibitors recommended |
| Cell culture supernatant | ≥ 20 mL (depending on cell type) | Harvest from conditioned medium; serum-free culture preferred |
| Other biofluids (CSF, saliva, etc.) | Contact us for specific requirements | Customized protocols available |
Additional Notes:
- Samples may be submitted as either pre-isolated exosomes or raw materials for exosome isolation by our team.
- Avoid high concentrations of salts, detergents, or ion-pairing reagents, which reduce MS sensitivity.
- Please consult our technical team before shipping for detailed packaging and storage instructions.
What Deliverables Will You Receive
- A comprehensive, publication-ready scientific report.
- A complete list of identified and quantified molecules (proteins, lipids, or metabolites) with statistical analysis.
- Raw data files (e.g., .raw) and processed data files.
- Detailed bioinformatics analysis, including PCA plots, volcano plots, heatmaps, and pathway enrichment analysis.
- A full summary of methods and all quality control data.
Applications of Mass Spectrometry in Exosome Research
Our mass spectrometry exosome services are widely applied in biomedical and translational research:
- Biomarker Discovery: Detection of disease-related proteins and lipids in exosomes from blood, urine, or other biofluids, aiding early diagnosis of cancers, neurodegenerative, cardiovascular, and metabolic diseases.
- Disease Mechanism Studies: Elucidation of exosome-mediated pathways in tumor metastasis, immune modulation, and inflammatory processes.
- Drug Delivery Evaluation: Assessment of exosomes as natural drug carriers, including drug loading, release, and in vivo stability.
- Exosome Product Quality Control: Verification of purity, stability, and molecular consistency in therapeutic or engineered exosome preparations.
Why Choose Creative Biostructure?
- Proven expertise in exosome proteomics, structural biology, and advanced MS technologies
- Comprehensive multi-omics profiling covering proteins, lipids, and metabolites
- High sensitivity and precision, including absolute quantification with stable isotope-labelled peptides
- Strict quality control and standardized reporting in line with MISEV2023 and MIAPE guidelines
- Trusted global partner with a dedicated scientific team supporting academic and industrial research
Case Study
Case: Proteomic and Metabolomic Profiling of T Cell-Derived Exosomes
Background
This study compared T cell-derived exosomes (CD3⁺) with non-T cell exosomes (CD3⁻) from human plasma using immune capture and mass spectrometry.
Methods
Exosomes from 10 donors were separated by size-exclusion chromatography and anti-CD3 capture. LC-MS/MS identified 418 proteins, while targeted metabolomics quantified 338 metabolites.
Results
CD3⁺ exosomes showed enrichment of 36 proteins and 96 metabolites, including cholesterol, sphingomyelins, ceramides, and hexoses, reflecting T cell biology. In contrast, CD3⁻ exosomes were linked to platelet and neutrophil activity.
Conclusion
Mass spectrometry revealed that CD3⁺ exosomes mirror T cell molecular features, supporting their use as a "liquid T cell biopsy" and highlighting the value of MS for immune monitoring and biomarker discovery.
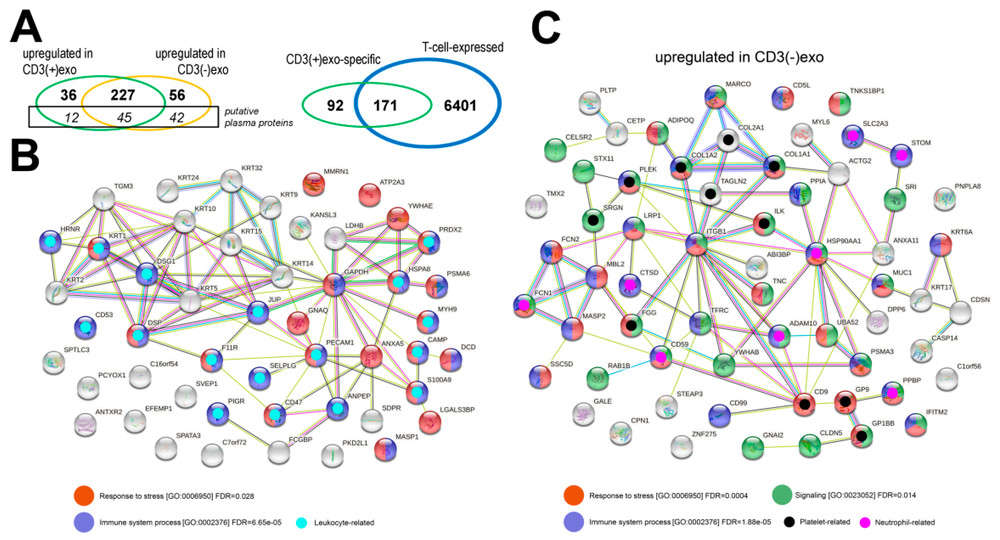 Figure 3. Proteomic Profiling of CD3(+) and CD3(-) Plasma sEV Fractions. (A, left) Venn diagram of proteins significantly upregulated (FDR < 5%) in CD3(+) vs. CD3(-) exosomes, with plasma proteins shown separately. (A, right) Overlap between CD3(+) exosome-specific proteins and known T cell proteins. (B) Functional protein networks enriched in CD3(+) exosomes. (C) Functional protein networks enriched in CD3(-) exosomes, with proteins color-coded by biological processes and annotated via STRINGdb. (Zebrowska A, et al., 2022)
Figure 3. Proteomic Profiling of CD3(+) and CD3(-) Plasma sEV Fractions. (A, left) Venn diagram of proteins significantly upregulated (FDR < 5%) in CD3(+) vs. CD3(-) exosomes, with plasma proteins shown separately. (A, right) Overlap between CD3(+) exosome-specific proteins and known T cell proteins. (B) Functional protein networks enriched in CD3(+) exosomes. (C) Functional protein networks enriched in CD3(-) exosomes, with proteins color-coded by biological processes and annotated via STRINGdb. (Zebrowska A, et al., 2022)
At Creative Biostructure, from biomarker discovery to therapeutic development, we deliver accurate, reproducible data tailored to your goals. Request a quote or contact us to discuss your project needs.
References
- Zebrowska A, Jelonek K, Mondal S, et al. Proteomic and metabolomic profiles of T cell-derived exosomes isolated from human plasma. Cells. 2022, 11(12): 1965.
- Welsh J A, Goberdhan D C I, O'Driscoll L, et al. Minimal information for studies of extracellular vesicles (MISEV2023): From basic to advanced approaches. Journal of Extracellular Vesicles. 2024, 13(2): e12404.
- Cheng X, Yu W, Liu Y, et al. Proteomic characterization of urinary exosomes with pancreatic cancer by phosphatidylserine imprinted polymer enrichment and mass spectrometry analysis. Journal of Proteome Research. 2024, 24(1): 111-120.
Frequently Asked Questions
For any inquiries, our support team is ready to help you get technical support for your research and maximize your experience with Creative Biostructure.