Integrated Exosome Proteomics Service with Single-Cell and Tissue Analysis
The high heterogeneity of cells and tissues presents a major bottleneck that hinders researchers from deeply deciphering the functional mechanisms of exosomes and discovering highly specific disease biomarkers. Creative Biostructure has innovatively introduced an integrated solution combining exosome proteomics, single-cell analysis, and tissue analysis. Our strategy effectively addresses the heterogeneity challenges in exosome research, significantly mitigating study biases caused by sample variability. Our specialized services provide robust evidence for cell tracing of exosomes and their regulatory mechanisms in disease, accelerating the discovery of clinically relevant biomarkers for our clients.
Why Integrate Exosomal Proteomics with Single-Cell and Tissue Analysis?
Exosomes are a type of extracellular vesicles, ranging in size from 50 to 150 nm, which carry various components (including nucleic acids, proteins, lipids, metabolites, and cell surface proteins) from their parent cells and shuttle through intercellular spaces to facilitate signal transmission in cell-to-cell communication. Since nearly all cell types secrete exosomes, those isolated from conventional research samples exhibit significant heterogeneity, including variations in size, cargo, function, and cellular origin.
Integrating exosome proteomics, single-cell omics, and tissue analysis technologies, this approach aims to explore the relationship between individual cells and their secreted exosomes within the tissue microenvironment. This cutting-edge service enables researchers to trace exosomes back to their specific cellular origins, decipher their protein cargo, and map their distribution and interactions in situ. This integrated analytical approach reveals cellular heterogeneity, exosomal communication mechanisms, and potential disease biomarkers with high resolution and multidimensional insight, providing robust support for precision medicine and drug development.
Our Integrated Exosome Proteomics Service with Single-Cell and Tissue Analysis
Creative Biostructure has developed a specialized single-cell combined exosome research service through extensive technical R&D. Clients provide a single tissue sample, which undergoes specialized composite collagenase digestion to separate tissue into single cells and interstitial fluid. Single-cell sequencing (scRNA-seq) of these cells yields cell subpopulation classification information within the tissue. Subsequently, proteomic analysis of exosomes in the interstitial fluid reveals the classification of tissue-derived exosome subpopulations. Integrating single-cell sequencing data with the exosome omics data enables identification of primary donor cells secreting tissue-derived exosomes or primary recipient cells internalizing them. This provides a crucial foundation for in-depth mechanistic studies and biomarker tracing.
Workflow of Integrated Exosome Proteomics Service with Single-Cell and Tissue Analysis
Our integrated service provides a seamless, end-to-end workflow designed to unlock a deeper understanding of cellular communication within the native tissue microenvironment. We guide your project from a single tissue sample to biologically actionable insights through a meticulously coordinated process.
Sample Consultation & Design
Receive sample types: Fresh/frozen tissue sections (10-20 μm), dissociated tissue cells, or tissue-derived exosome suspensions.
Collaborate with clients to define research goals (e.g., "Identify exosome sources in breast cancer tissue") and select modules (e.g., scRNA-seq + exosome proteomics + mIF).
Sample Processing & Multi-Omics Detection
Tissue preparation: FFPE or fresh frozen tissue sectioning, LCM for region-specific isolation.
Parallel analyses
Single-cell omics: scRNA-seq/CyTOF of dissociated tissue cells.
Exosome analysis: Isolation + MS-based proteomics of tissue-region exosomes.
Spatial omics: Visium spatial transcriptomics or mIF of tissue sections.
Data Integration & Analysis
Use custom bioinformatics pipelines (R, Python, Seurat, SpatialData) to unify multi-omics data.
Generate visualizations (spatial heatmaps, interaction networks, volcano plots) and statistical validation (P < 0.05, FC ≥ 1.2).
Report Delivery & Post-Service Support
Deliver comprehensive report: Experimental methods, QC data (e.g., NTA/TEM images, scRNA-seq cell clustering), integrated analysis results, and biological conclusions.
Provide 1:1 data interpretation meetings with multi-omics experts.
Assist with manuscript preparation (methodology sections, figure legends).
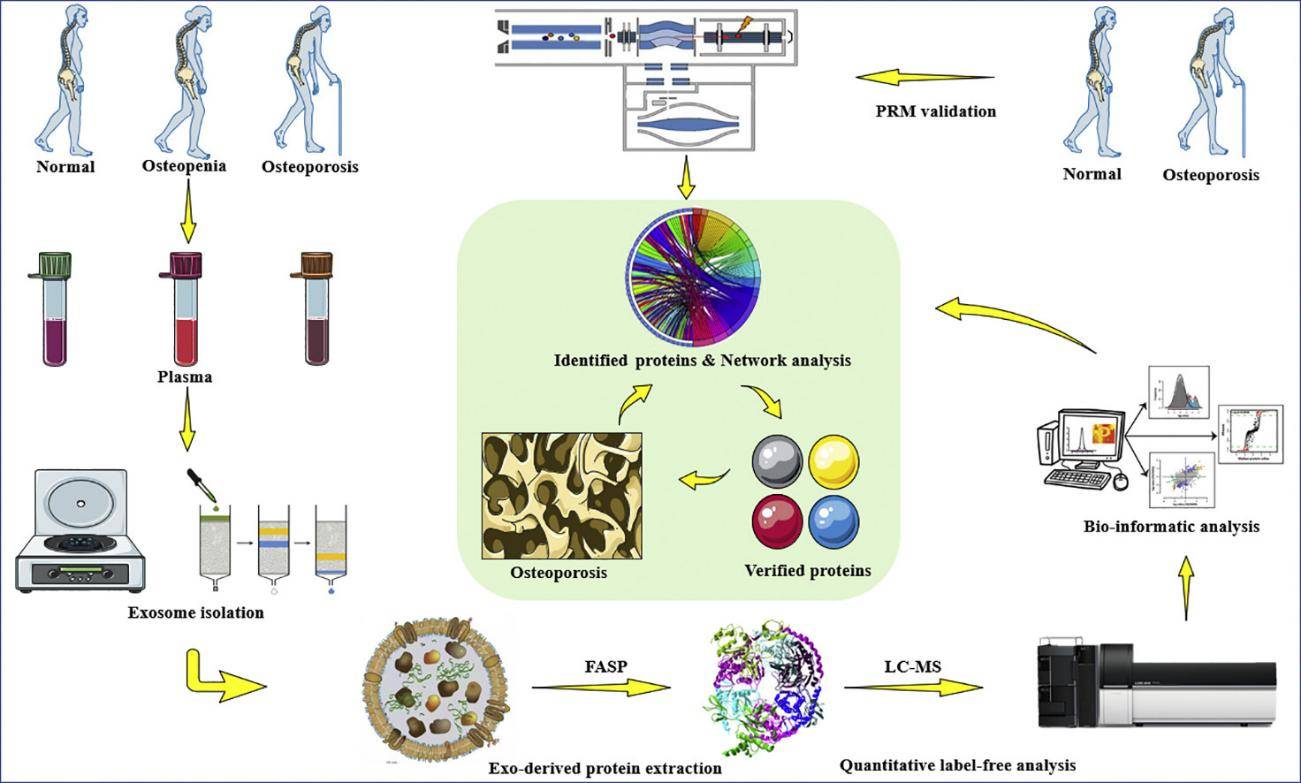
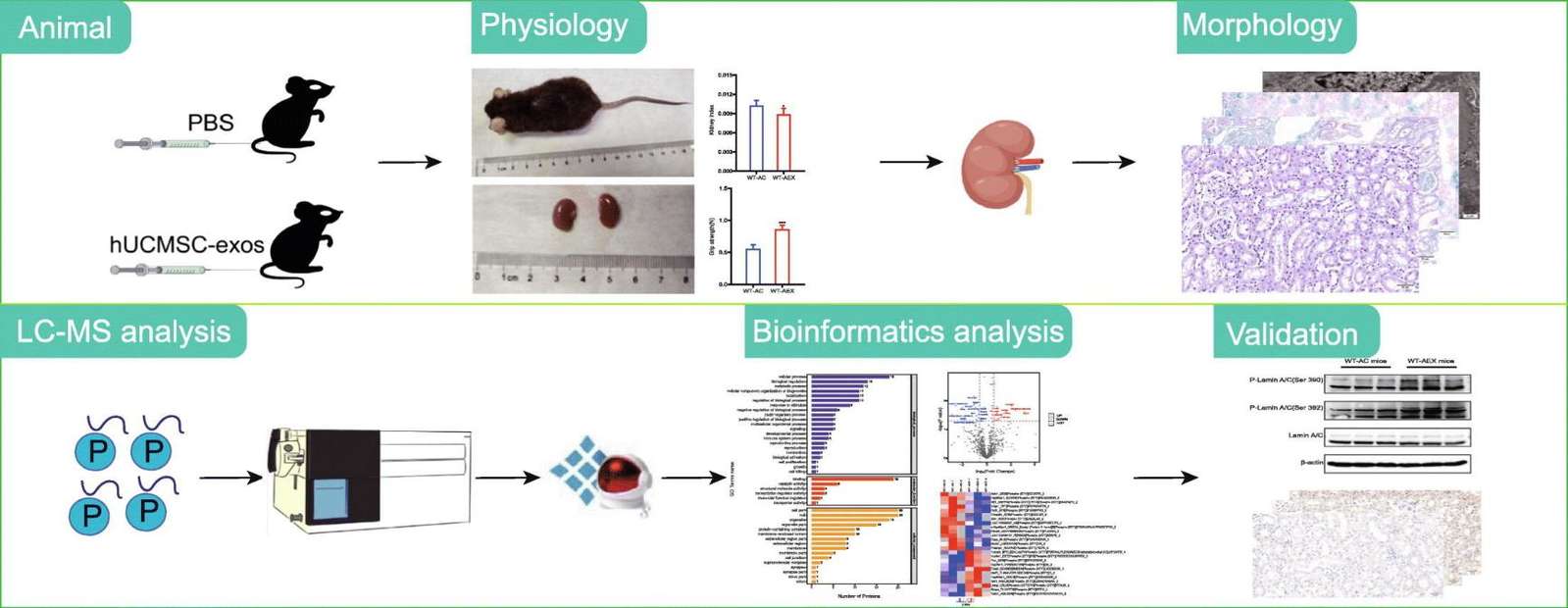
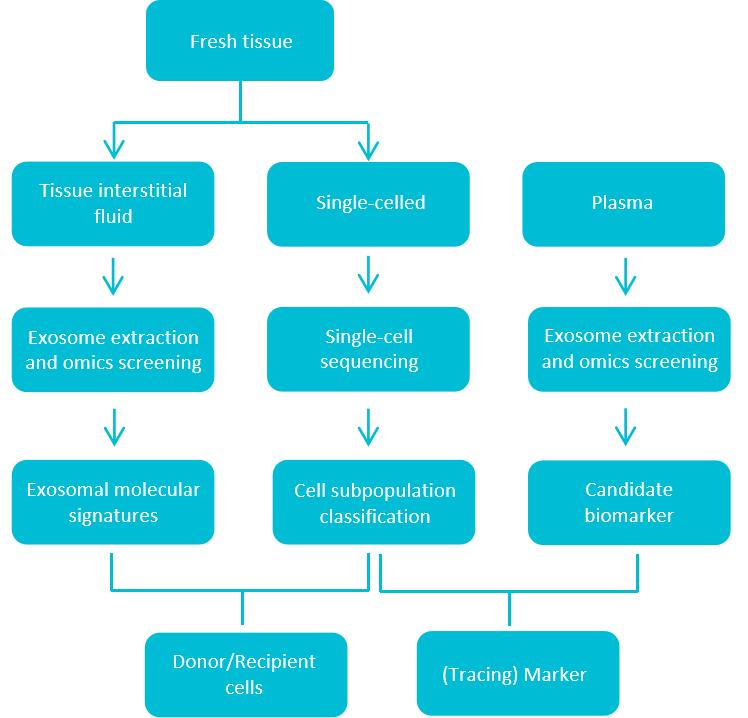 Figure 1. Workflow of integrated exosome proteomics service with single-cell and tissue analysis. (Creative Biostructure)
Figure 1. Workflow of integrated exosome proteomics service with single-cell and tissue analysis. (Creative Biostructure)
Deliverables
- Raw data: scRNA-seq FASTQ files, exosome MS .raw files, spatial omics count matrices.
- Processed data: Cell clustering results, exosome protein lists (with quantification), spatial maps.
- Analysis report: Integrated conclusions, statistical tables, and high-resolution visualizations.
- Technical documentation: Sample processing protocols, QC certificates, and software parameters.
Case Study
Case: Multi-omics Profiling of Serum EVs Identifies MACROH2A1 in Refractory COVID-19.
Background
To identify potential serum biomarkers and examine proteins associated with the pathogenesis of refractory COVID-19, this study performed high-coverage proteomic analysis on serum extracellular vesicles collected from 12 COVID-19 patients with varying disease severity and 4 healthy controls. Additionally, single-cell RNA sequencing was conducted on peripheral blood mononuclear cells (PBMCs) obtained from 10 COVID-19 patients and 5 healthy controls.
Methods
- Isolation of EVs for proteomics.
- Proteomic analysis.
- Bioinformatics analysis of the proteome.
- Analysis of molecular networks and pathways for the proteome and transcriptome.
- Cell culture and stimulation.
- Transmission electron microscopy.
- Subjects and specimen collection of PBMCs for scRNA-seq.
- Droplet-based single-cell sequencing.
- Alignment, quantification, and quality control of scRNA-seq data
Conclusion
The study demonstrates that MACROH2A1 in extracellular vesicles serves as a potential biomarker for refractory COVID-19, possibly reflecting the disease pathogenesis in monocytes.
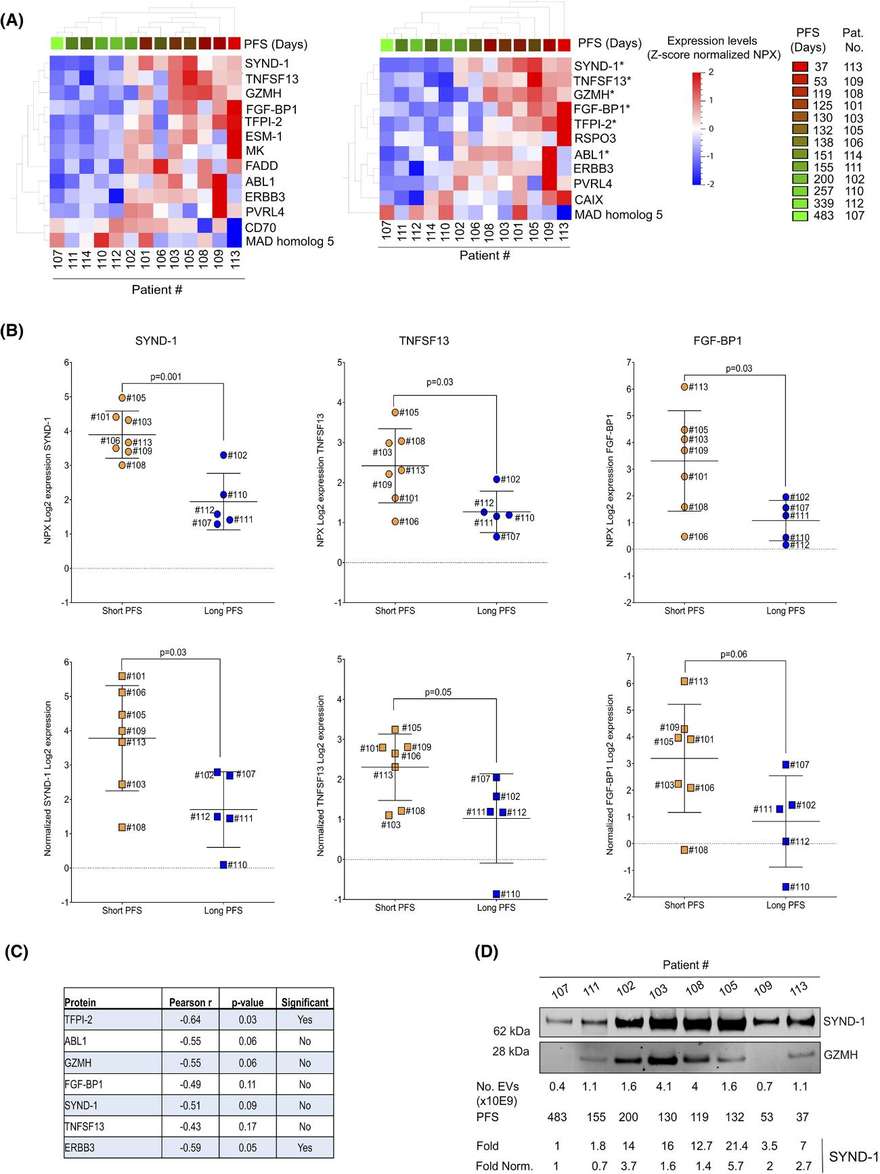
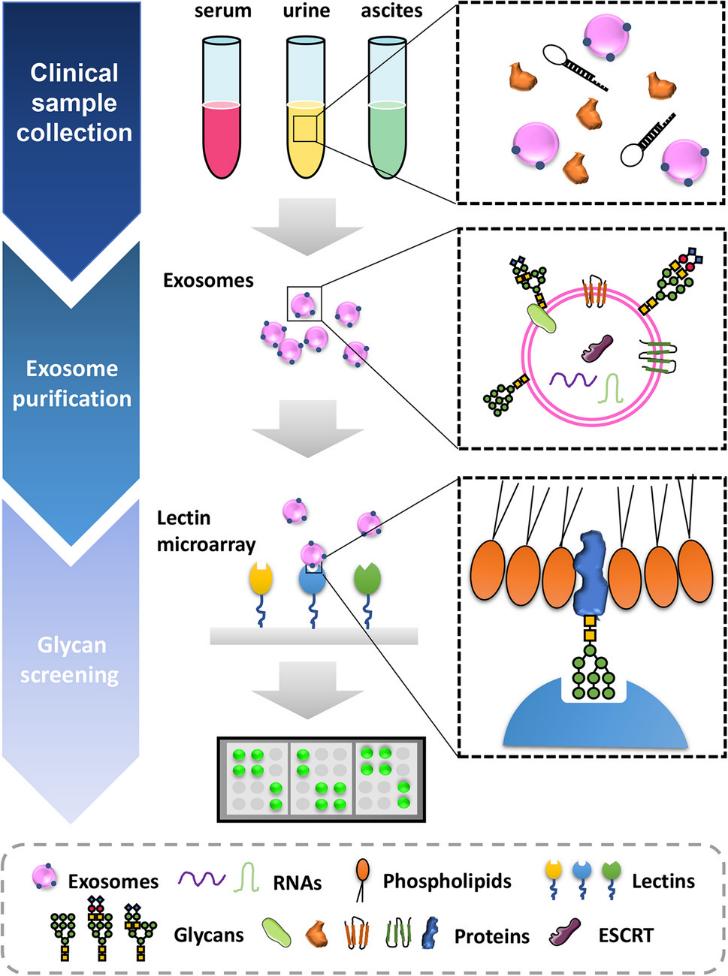
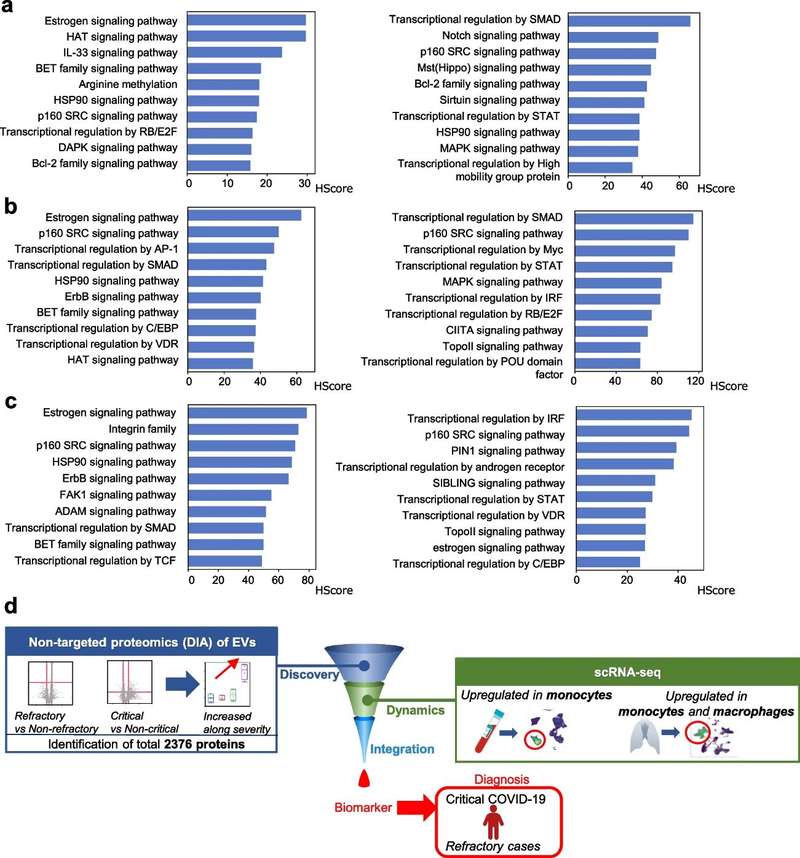 Figure 2. Next-generation proteomics of serum extracellular vesicles combined with single-cell RNA sequencing identifies MACROH2A1 associated with refractory COVID-19. (Kawasaki T, et al., 2022)
Figure 2. Next-generation proteomics of serum extracellular vesicles combined with single-cell RNA sequencing identifies MACROH2A1 associated with refractory COVID-19. (Kawasaki T, et al., 2022)
Ready to turn tissue heterogeneity from noise into insight? Our integrated exosome proteomics, single-cell, and tissue analysis workflow traces vesicles back to their cellular origins, maps their cargo in situ, and links them to clinically relevant phenotypes with multi-omics, publication-ready reports. Contact us to tailor your study design and receive a fast, expert quotation.
Reference
- Kawasaki T, Takeda Y, Edahiro R, et al. Next-generation proteomics of serum extracellular vesicles combined with single-cell RNA sequencing identifies MACROH2A1 associated with refractory COVID-19. Inflammation and Regeneration. 2022, 42(1): 53.
Frequently Asked Questions
For any inquiries, our support team is ready to help you get technical support for your research and maximize your experience with Creative Biostructure.