Exosome Quantitative Proteomics Services
Exosome proteomics is a rapidly evolving research field that focuses on high-efficiency quantification and analysis of proteins within exosomes. Creative Biostructure provides a comprehensive technical service system for exosome proteomics research, covering projects from exosome collection and characterization to qualitative/quantitative proteomic profiling and data analysis. Our integrated approach ensures the generation of high-quality, reliable research data under consistent technical standards.
Why is Exosome Quantitative Proteomics Important?
Exosome quantitative proteomics is critically important because it provides an unbiased, high-resolution view of the protein cargo that exosomes carry, which directly reflects the functional state and specific activities of their parent cells. By precisely measuring protein abundances, this powerful approach enables the discovery of novel biomarkers for early disease diagnosis, reveals new insights into disease mechanisms, and identifies potential therapeutic targets. As a non-invasive liquid biopsy strategy, it is particularly valuable for monitoring treatment response and disease progression in complex conditions like cancer and neurodegenerative disorders, thereby bridging fundamental biological discovery with clinical translation.
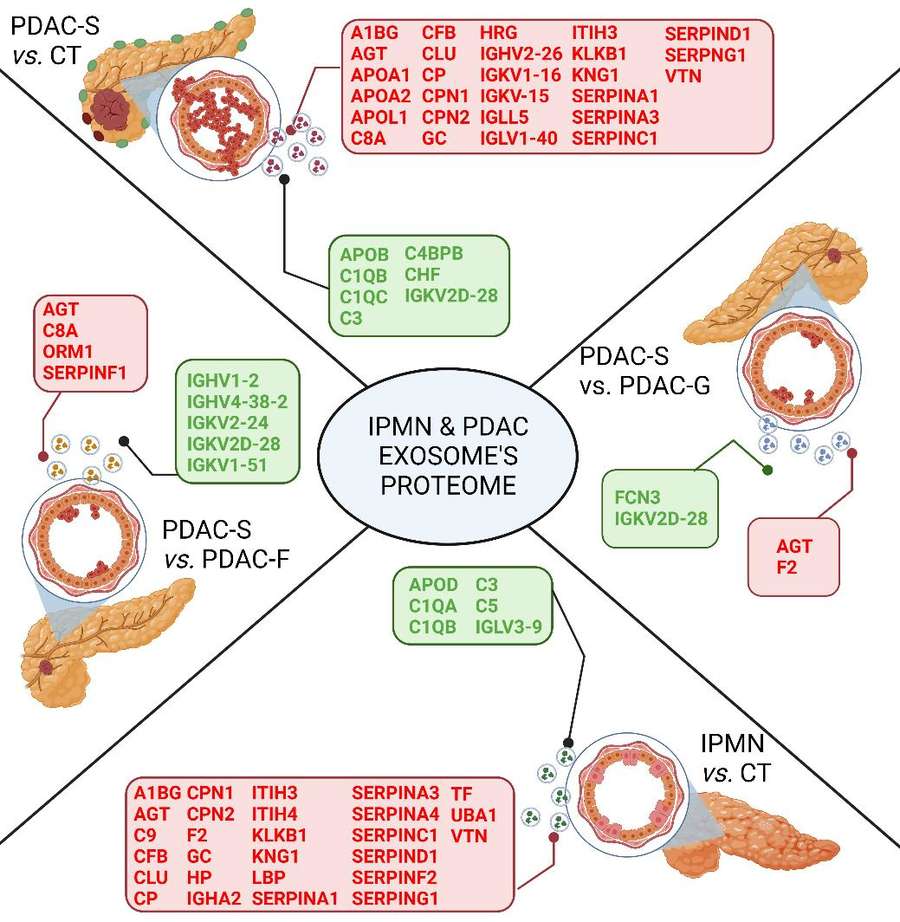
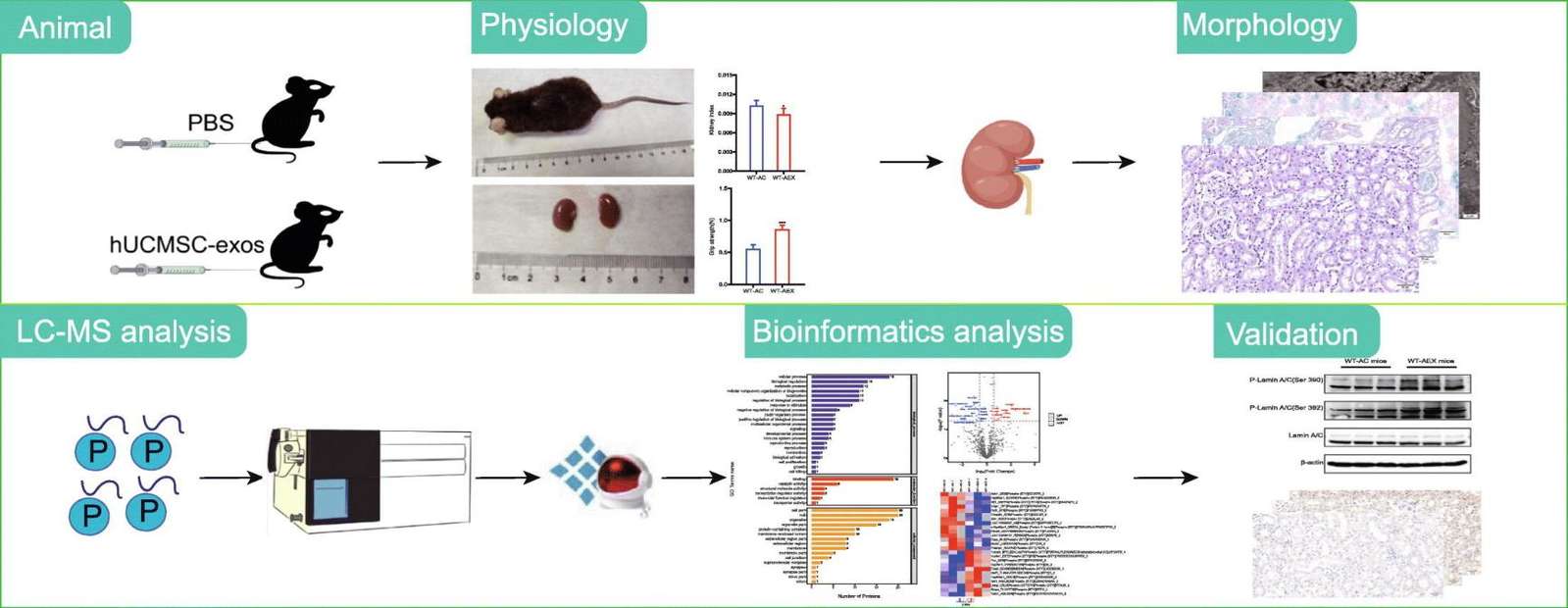
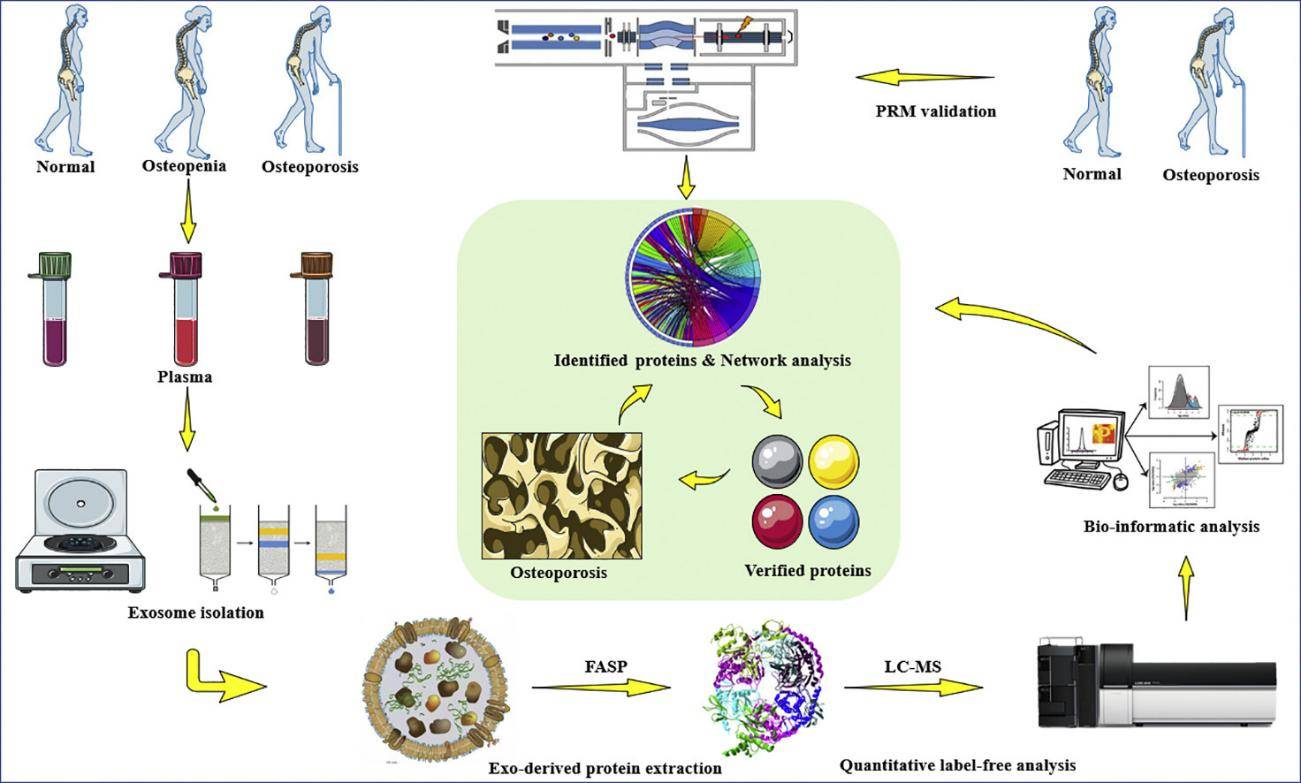 Figure 1. Plasma exosome protein markers in osteoporosis identified by quantitative proteomics and reverse engineering. (Chen M, et al., 2020)
Figure 1. Plasma exosome protein markers in osteoporosis identified by quantitative proteomics and reverse engineering. (Chen M, et al., 2020)
Our Technical Platform for Exosome Quantitative Proteomics
Our exosome quantitative proteomics service is built on a core technical platform relying on high-precision mass spectrometry systems, including advanced instruments such as Orbitrap Eclipse and Q Exactive HF-X, which can achieve high-resolution separation and accurate quantification of peptides. Auxiliary technical equipment includes:
| Platform | Description |
|---|---|
| Exosome isolation system | Ultra-high-speed centrifuge (150,000×g) and automatic exosome purification instrument, supporting standardized processing of batch samples |
| Multi-dimensional separation system | Two-dimensional liquid chromatography (2D-LC) combined with reversed-phase chromatography technology to improve the separation efficiency of complex protein mixtures |
| Quantitative labeling platform | Equipped with iTRAQ/TMT labeling reagents and Label-free quantitative analysis system to meet quantitative needs of different throughputs. |
| Data analysis system | High-performance computing cluster supports large-scale data processing of software such as MaxQuant and Perseus, realizing integrated analysis of protein quantification and functional annotation. |
Exosome Quantitative Proteomics Analysis Workflow
Creative Biostructure maintains a specialized technical team. We will thoroughly understand your research context, sample types (e.g., serum/plasma, cell supernatant), experimental objectives (e.g., disease biomarker screening, drug efficacy monitoring), and requirements for quantification accuracy and detection throughput. Based on this information, we provide tailor-made experimental proposals, including recommendations for optimal quantification strategies (iTRAQ/TMT labeling or Label-free quantification), sample processing methods, instrument specifications, and data analysis components, along with detailed quotations and project timeline projections.
Our standard workflow includes the following key steps:
Sample Receipt and Quality Control
After receiving the samples, information verification is completed promptly. A sample quality control report is generated through appearance inspection, concentration determination (BCA method), and integrity evaluation (such as hemolysis detection for serum samples).
Exosome Isolation and Purification
The combined technology of "ultra-high-speed centrifugation + size exclusion chromatography (SEC)" is adopted: exosomes are first enriched by centrifugation at 120,000×g for 2 hours, then protein aggregates are removed through the SEC column, and finally high-purity exosomes are obtained (verified by marker protein WB).
Protein Extraction and Processing
Total proteins are extracted by RIPA lysate containing protease inhibitors, and then enzymatically digested by FASP (trypsin at 37℃ for 16 h) to produce peptides, which were desalted on a C18 column and then partitioned into samples for labeled quantitation (iTRAQ/TMT) or unlabeled quantitation (Label-free), respectively.
Mass Spectrometry
The peptide segments are separated by liquid chromatography and subsequently introduced into an Orbitrap mass spectrometer for data acquisition.
- Labeling quantification: TMT 10-plex labeling is used, with a resolution of 60,000 for primary mass spectra and 15,000 for secondary mass spectra.
- Non-labeled quantification: DDA mode was adopted, 20 secondary spectra were collected in each cycle to ensure the quantitative accuracy of CV<10%.
Data Analysis
The raw data are matched with UniProt database by MaxQuant software to complete protein identification and quantitative analysis, and then analyzed by GO/KEGG enrichment analysis and protein-protein interaction network (PPI) construction to analyze the biological functions.
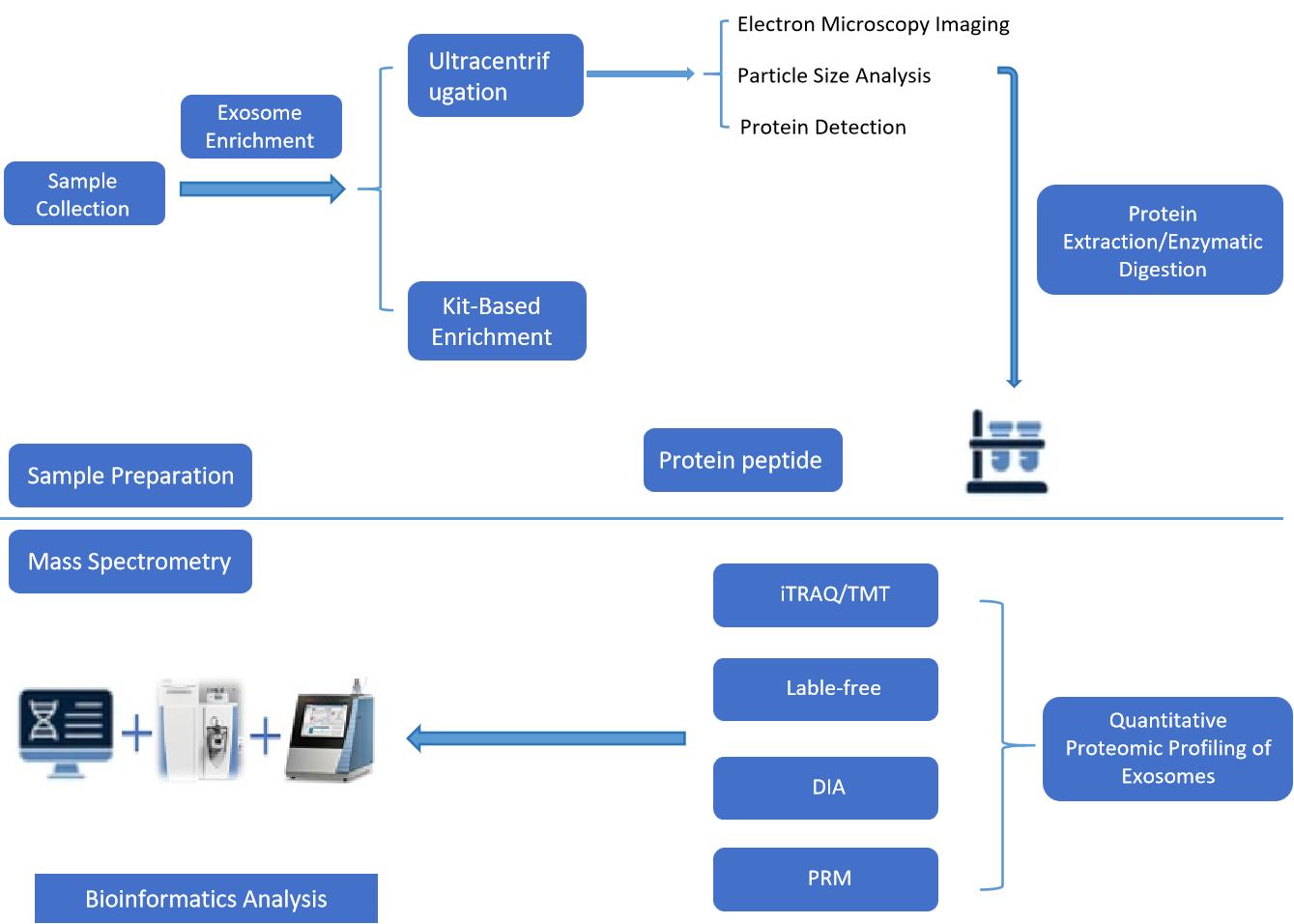 Figure 2. Standard workflow of exosome quantitative proteomics service. (Creative Biostructure)
Figure 2. Standard workflow of exosome quantitative proteomics service. (Creative Biostructure)
Supported Sample Range
To meet your diverse research needs, our service supports the analysis of the following sample types:
| Sample Type | Minimum Requirement | Recommended Storage Condition |
|---|---|---|
| Serum/Plasma | 1mL | -80℃, EDTA anticoagulation |
| Cell Supernatant | 10mL | -80℃, serum-free medium |
| Urine | 20mL | 4℃ with preservatives added |
| Cerebrospinal Fluid/Pleural Effusion | 5mL | -80℃, avoid repeated freezing and thawing |
| Purified Exosomes | 1×10^10 particles | -80℃ suspended in PBS |
Deliverables
Experimental Documentation
- Sample processing records (including QC reports, exosome characterization profiles).
- Mass spectrometry parameter settings and raw spectral data (raw format).
Data Files
- Protein identification list (including peptide sequence, coverage, molecular weight, etc.).
- Quantitative results matrix (differential protein screening criteria: FC ≥ 1.2, P < 0.05).
- Visualization charts (volcano, cluster heatmap, pathway-enriched bubble map).
Analysis Report
- Detailed description of technical methods and data quality assessment.
- Functional interpretation of differential proteins and recommendation of potential markers.
- Suggestions for design of follow-up validation experiments (e.g., PRM-targeted quantification).
Applications of Our Exosome Quantitative Proteomics Services
Screening of Disease Biomarkers
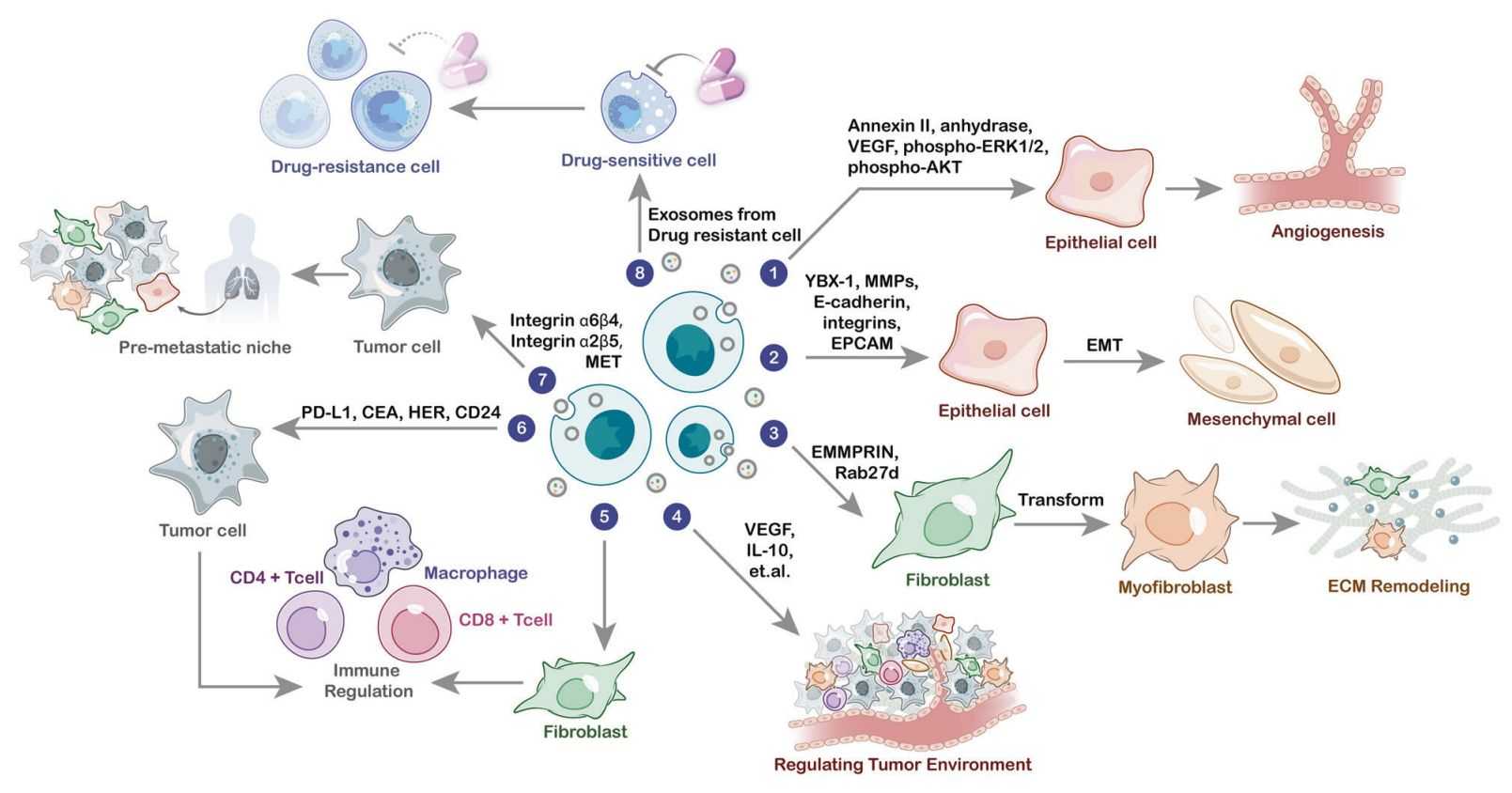
Compare the differences in serum exosomal proteins between cancer patients and healthy individuals, and screen diagnostic biomarkers using machine learning models (e.g., combined detection of CEA and CA19-9 in exosomes from colorectal cancer patients).
Monitoring of Drug Efficacy
Analyze changes in exosomal proteins in tumor patients before and after chemotherapy. For example, the decreased level of p-AKT protein in exosomes of breast cancer patients after paclitaxel treatment can serve as an indicator for efficacy evaluation.
Mechanistic Research
Elucidate cell communication mediated by exosomal proteins, such as how TGF-β1 in exosomes secreted by cancer-associated fibroblasts (CAFs) promotes epithelial-mesenchymal transition (EMT) in cancer cells.
Advantages of Our Exosome Quantitative Proteomics Services
- Highly Accurate Quantification: Using TMT 16-plex labeling technology, simultaneous quantification of multiple sample groups can be achieved, reducing inter-batch variability and ensuring result reproducibility.
- Deep Coverage: Our high-sensitivity platform achieves deep proteome coverage of exosomes, enabling sensitive detection of low-abundance functional proteins (e.g., cytokines, receptor proteins) across the full-scale molecular weight range.
- Full-Process Quality Control: Strict quality control standards are set for key steps such as exosome purity, protein extraction efficiency, and peptide recovery rate to ensure accurate testing data for customers.
- Customized Analysis: We support personalized needs, such as phosphorylated protein quantification, protein-protein interaction network analysis, and multi-omics integration analysis (integration with transcriptome/metabolome data).
Case Study
Case: CML Imatinib Resistance: Plasma Exosome Proteomic Biomarker Discovery
Background
Exosome-harbored proteins were involved in tumor drug resistance and could be novel biomarkers for the diagnosis and drug sensitivity prediction of cancer. The aim of this study was to investigate the proteomic profile of plasma exosomes derived from CML patients to identify ideal biomarkers for IM resistance.
Methods
Exosomes were extracted via ultracentrifugation from pooled plasma samples of 9 imatinib-resistant and 9 imatinib-sensitive chronic myeloid leukemia (CML) patients. Expression levels of exosomal proteins were identified using label-free quantification based on liquid chromatography-tandem mass spectrometry (LC-MS/MS). Proteomic data were analyzed through bioinformatics methods, and candidate proteins were validated by western blot (WB) and parallel reaction monitoring (PRM) analyses.
Conclusion
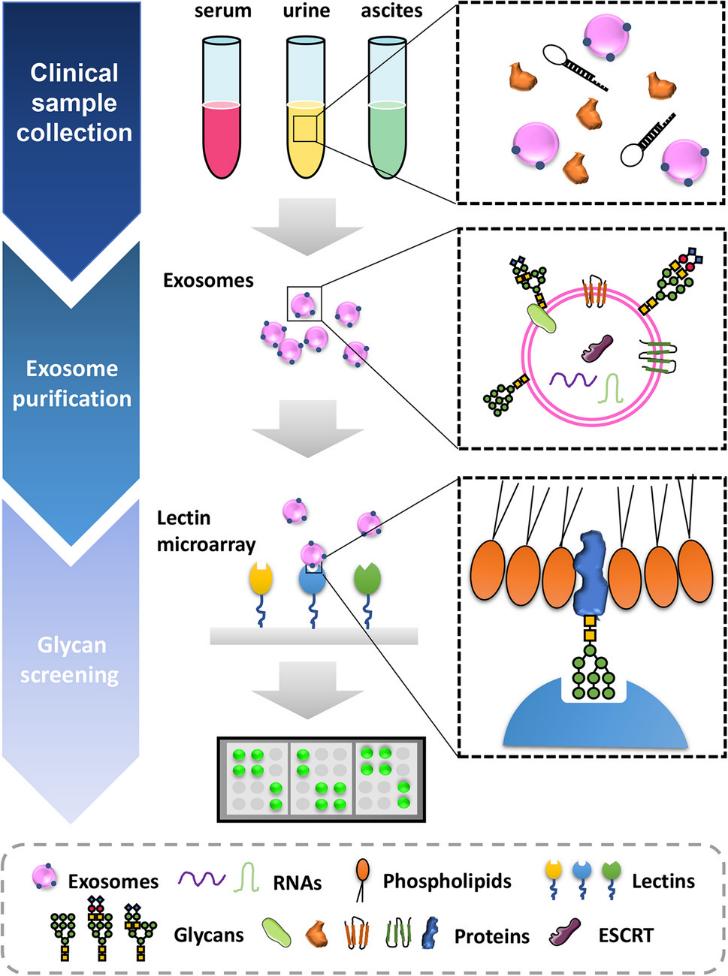
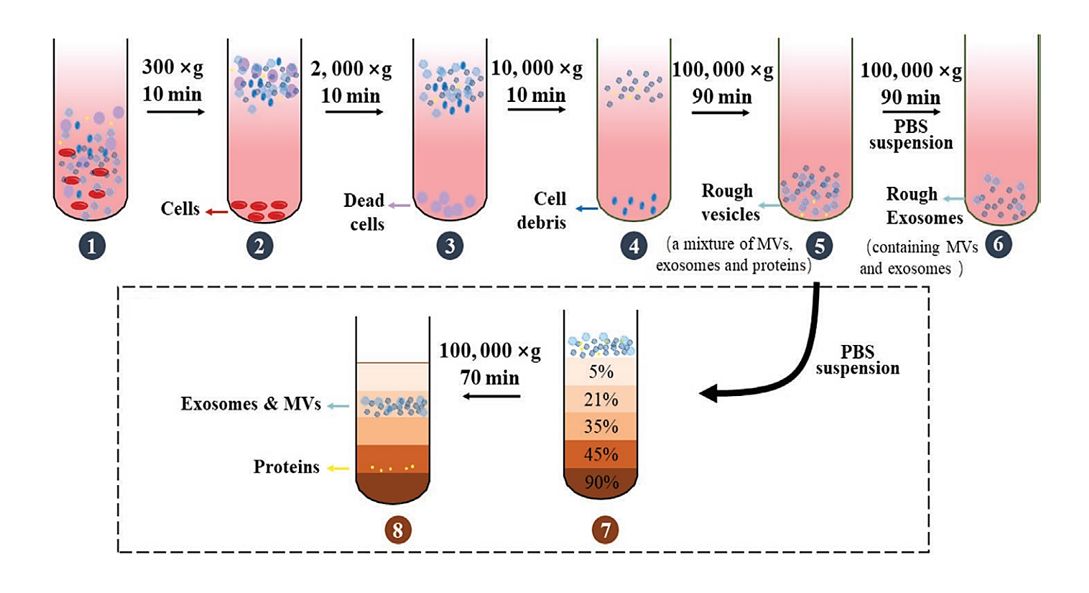
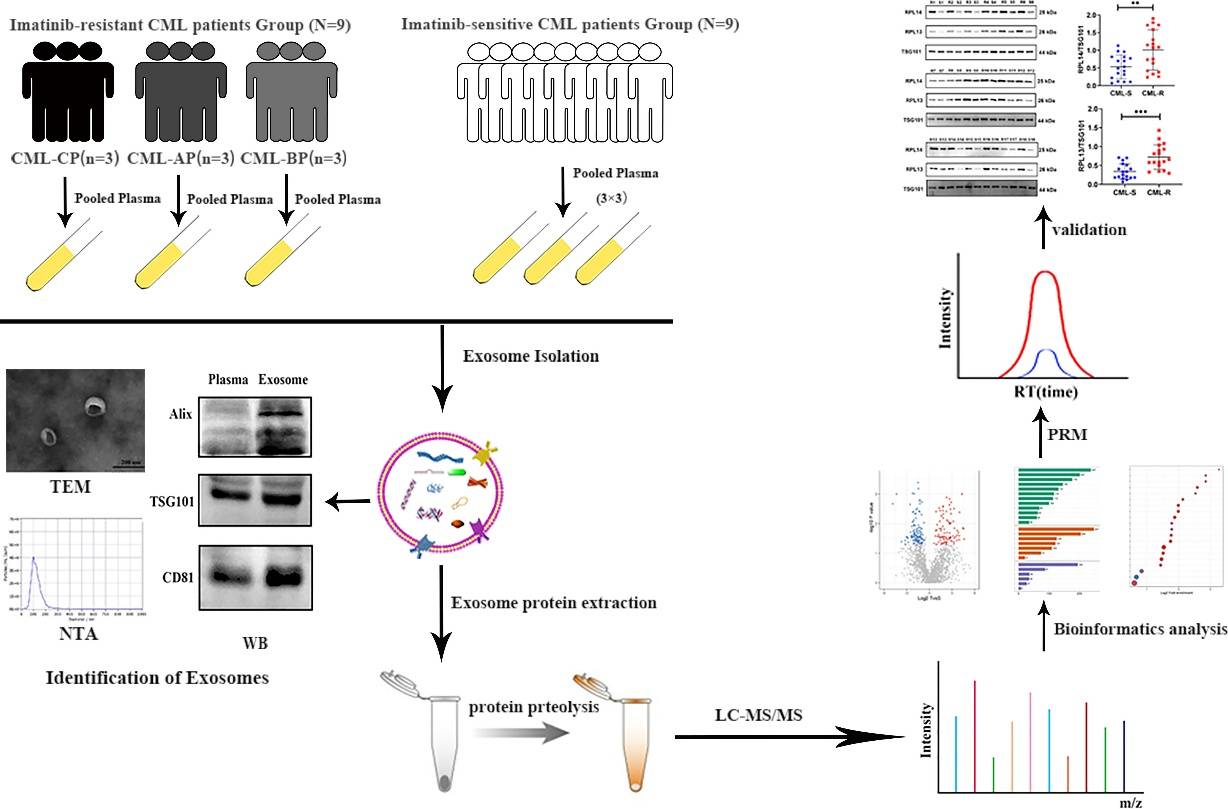 Figure 3. Schematic workflow shown the LC-MS/MS-based quantitative proteomic analysis of exosome isolated from plasma samples of IM-R and IM-S CML patients. (Li M Y, et al., 2021)
Figure 3. Schematic workflow shown the LC-MS/MS-based quantitative proteomic analysis of exosome isolated from plasma samples of IM-R and IM-S CML patients. (Li M Y, et al., 2021)
Proteomic analysis of plasma exosomes provides new ideas and important information for the study of IM resistance in CML. Especially the exosomal proteins (RPL13 and RPL14), which may have great potential as biomarkers of IM resistance.
Creative Biostructure's exosome quantitative proteomics service centers on high-accuracy mass spectrometry detection, encompassing the complete workflow from sample reception, exosome isolation/purification, to protein quantification and data interpretation. Whether exploring molecular mechanisms of disease pathogenesis or developing novel diagnostic biomarkers, we deliver robust technical support to propel your research breakthroughs. Contact us today to discuss your project and receive a personalized solution.
References
- Chen M, Li Y, Lv H, et al. Quantitative proteomics and reverse engineer analysis identified plasma exosome derived protein markers related to osteoporosis. Journal of Proteomics. 2020, 228: 103940.
- Li M Y, Zhao C, Chen L, et al. Quantitative proteomic analysis of plasma exosomes to identify the candidate biomarker of imatinib resistance in chronic myeloid leukemia patients. Frontiers in oncology. 2021, 11: 779567.
Frequently Asked Questions
For any inquiries, our support team is ready to help you get technical support for your research and maximize your experience with Creative Biostructure.