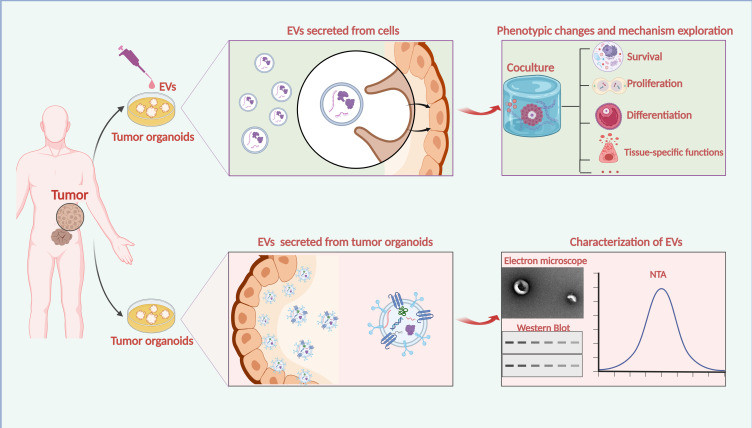
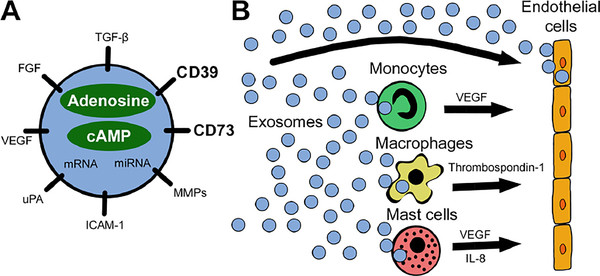
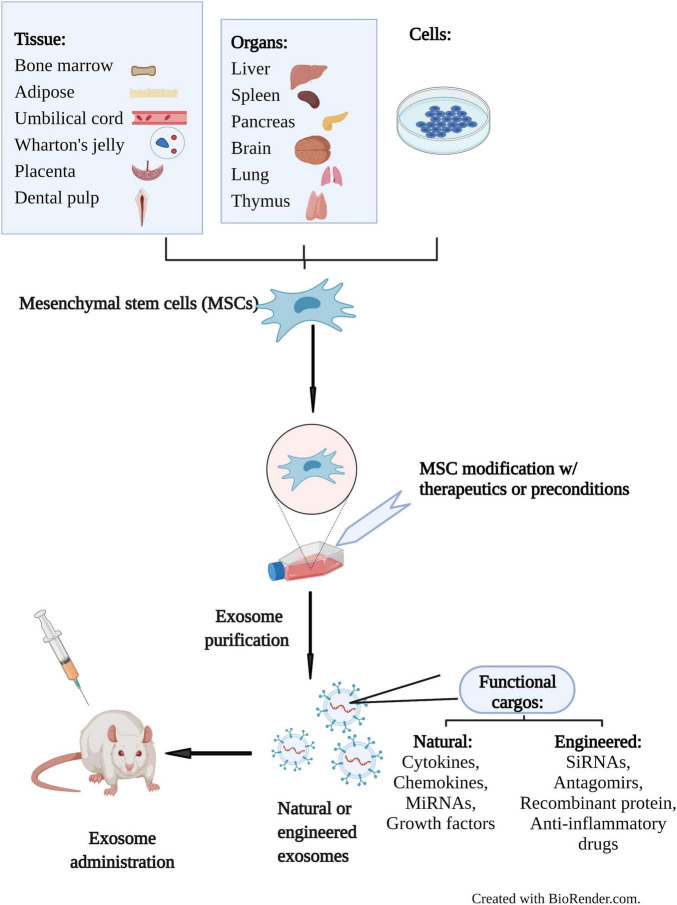
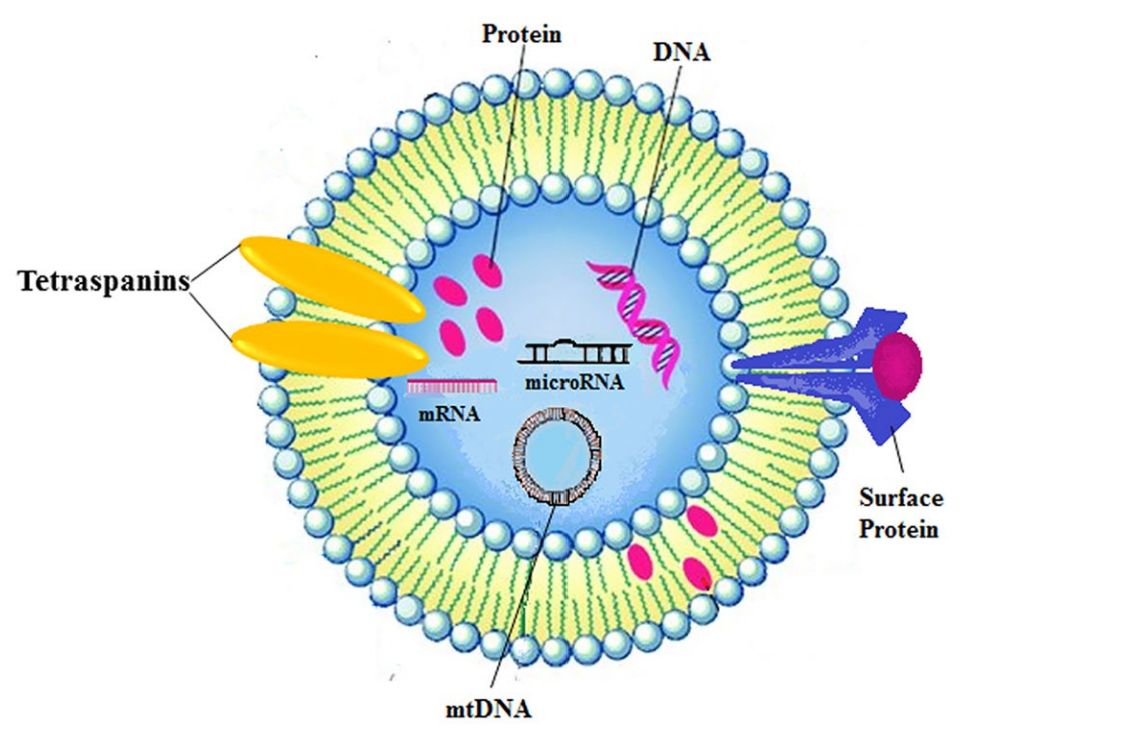
Exosome Dual-luciferase Reporter Gene Assay
Exosomes are natural carriers capable of transporting functional nucleic acids like microRNAs (miRNAs) and siRNAs, which play critical roles in regulating gene expression. While sequencing can identify the presence of these molecules, it cannot confirm their biological activity. The crucial next step is to obtain definitive proof: does an exosome-delivered miRNA directly engage and suppress its intended target gene within the recipient cell?
Our Exosome Dual-luciferase Reporter Gene Assay is the gold-standard method to quantitatively answer this question. By cloning a target gene's 3' UTR sequence downstream of a Firefly luciferase reporter, this assay provides a direct readout of miRNA activity. A decrease in the luciferase signal upon exosome treatment serves as conclusive, quantitative evidence of functional nucleic acid delivery and target engagement.
Why Use a Dual-Luciferase Assay for Exosome Cargo Validation?
While sequencing can tell you what RNA is inside your exosomes, it cannot confirm biological function. The dual-luciferase system offers unparalleled advantages:
- Direct Functional Readout: Directly measures target gene repression (for miRNA/siRNA) or protein expression (for mRNA) mediated by the exosome cargo.
- Quantitative & Sensitive: Provides numerical data on the degree of regulation, sensitive enough to detect subtle effects.
- Internal Normalization: The "dual" system uses a second reporter (Renilla luciferase) to normalize for variations in cell number and transfection efficiency, ensuring highly reliable and reproducible results.
- Gold Standard: Widely accepted as the definitive method for validating miRNA target interactions and siRNA knockdown efficiency.
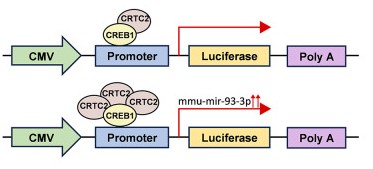 Figure 1. The pmirGLO vector contained Firefly luciferase and Renilla luciferase, and Renilla luciferase was used as a control. A schematic diagram shows the structure of the luciferase reporter plasmid. Luciferase activity was enhanced when the CREB1/CRTC2 complex was increased. (Sun Y, et al., 2025)
Figure 1. The pmirGLO vector contained Firefly luciferase and Renilla luciferase, and Renilla luciferase was used as a control. A schematic diagram shows the structure of the luciferase reporter plasmid. Luciferase activity was enhanced when the CREB1/CRTC2 complex was increased. (Sun Y, et al., 2025)
Our Dual-Luciferase Reporter Assay Platform
We provide an end-to-end service, from custom vector design and validation to final data analysis, specifically tailored for exosome-mediated nucleic acid delivery. Our platform combines state-of-the-art molecular biology with high-sensitivity detection to deliver definitive proof of your nucleic acid's bioactivity.
Key Research Services Offered
Our platform is designed to provide definitive answers to several critical research questions:
- miRNA Target Validation Service: We can conclusively prove that an exosome-delivered miRNA directly binds to and represses its predicted mRNA target, validating a key biological mechanism.
- siRNA Knockdown Efficiency Service: We can precisely quantify the functional knockdown efficiency of your exosome-delivered therapeutic siRNA, providing essential potency data.
- Functional mRNA Delivery Service: We can confirm that an exosome-delivered mRNA is successfully translated within the recipient cell and is biologically active.
- Comparative Potency Analysis: We can directly compare the nucleic acid delivery efficiency of exosomes from different sources or with different engineering modifications.
Our Technical Approach
Our technical approach is built on the highly reliable dual-luciferase system. We link your target sequence (e.g., a gene's 3' UTR) to a Firefly luciferase reporter gene. If your exosome delivers a functional miRNA or siRNA, it will bind this target and suppress the Firefly light signal. To prove specificity, we run a crucial control using a mutated target sequence that your cargo cannot bind. For accuracy, a second, constant Renilla luciferase is used to normalize the data, correcting for any experimental variations. This robust design provides clear, quantitative proof of functional cargo delivery.
Service Highlights and Advantages
- Custom Vector Construction: We handle the entire process of designing and cloning your specific WT and MUT target sequences into high-performance vectors.
- Flexible Cell Line Options: We can establish the reporter system in standard, easy-to-transfect cell lines (e.g., HEK293T) or in your specific target cell line to ensure physiological relevance.
- High-Sensitivity Detection: We utilize state-of-the-art luminometers with dual injectors, capable of capturing rapid and sensitive light signals with a wide dynamic range, ensuring accurate quantification.
- End-to-End Scientific Support: Our PhD-level scientists provide expert consultation throughout the project, from initial experimental design to the final interpretation of your results.
Our Assay Workflow
Our end-to-end workflow is meticulously designed to provide definitive proof of your nucleic acid's bioactivity. The entire service process is built around several critical stages to ensure accuracy and reliability.
Key Stages of Our Service
Assay System Design & Construction
This foundational stage involves designing and building the specific tools for your experiment. We custom-clone your precise target sequence (e.g., a gene's 3' UTR) into our Firefly luciferase reporter vectors, creating both Wild-Type and Mutant versions for specificity testing.
Reporter System Validation
Before testing your exosomes, we first establish the reporter cell line by transfecting the vectors into the chosen cells. We then validate the system's responsiveness to ensure it provides a sensitive and reliable "testing ground."
Exosome Functional Assessment
This is the core experimental phase. We treat the validated reporter cells with your exosome preparations alongside all necessary controls. After an optimized incubation period, we perform the high-sensitivity dual-luciferase measurement.
Our 5-Step Assay Pathway
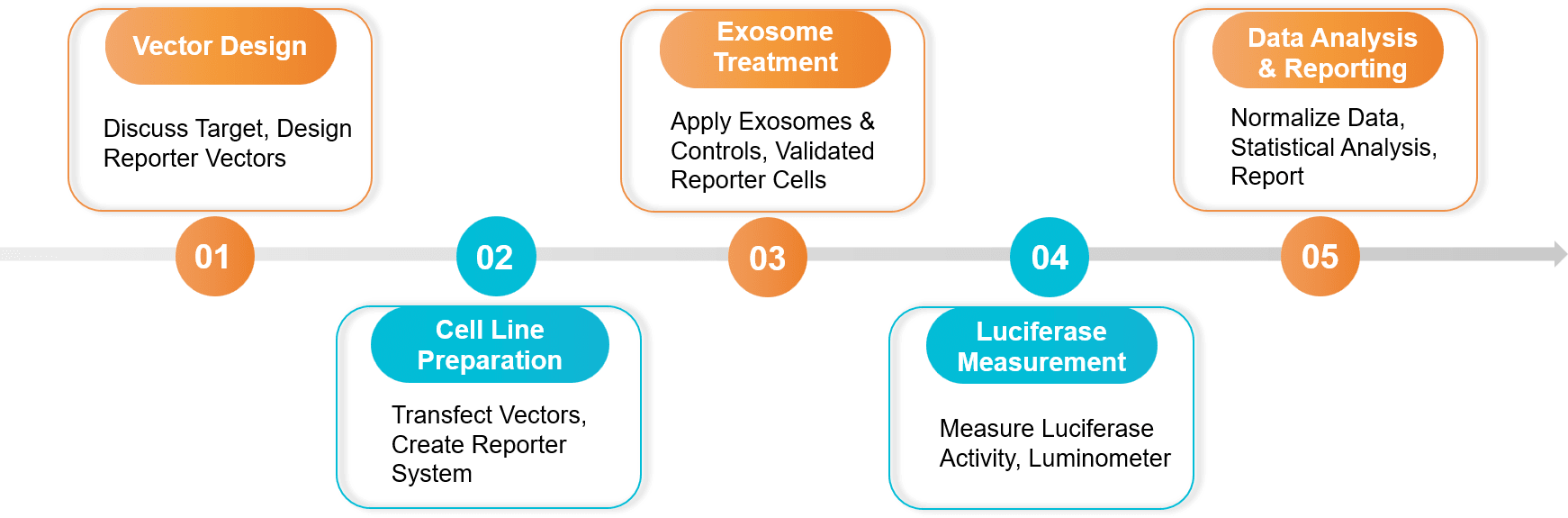 Figure 2. Exosome Dual-luciferase Reporter Gene Assay Project Workflow. (Creative Biostructure)
Figure 2. Exosome Dual-luciferase Reporter Gene Assay Project Workflow. (Creative Biostructure)
Sample Requirements
- Exosome Test Article:
- Purified exosome solutions containing the miRNA, siRNA, or mRNA of interest.
- Characterization data (NTA, cargo sequencing/qPCR if available) is highly recommended.
- Sequence Information:
- The specific sequence of the miRNA/siRNA/mRNA cargo.
- The identity and sequence of the predicted target gene/site for reporter vector construction.
- Recipient Cell Line: We can use standard lines (e.g., HEK293T, HeLa) or your specific target cell line.
Standard Deliverables
- A comprehensive report detailing vector design, experimental setup, and protocols.
- Raw luminescence data (RLU - Relative Light Units) for both Firefly and Renilla reporters.
- Normalized Firefly/Renilla ratios for all samples.
- Statistical analysis and graphical representation of the results (e.g., bar charts showing % repression).
- Scientific interpretation of the functional cargo delivery efficacy.
Case Study
Case: Proving Exosome-Delivered miR-210 Directly Targets the Apoptotic Gene AIFM3
Background: Researchers observed that exosomes from hypoxia-challenged mesenchymal stem cells (MSCs) could protect heart cells from apoptosis after a heart attack. They suspected this effect was due to the exosome-delivered cargo, specifically microRNA-210 (miR-210). Their hypothesis was that miR-210 directly suppresses a pro-apoptotic gene, AIFM3. They needed definitive molecular proof of this target engagement.
Methodology: To prove this direct molecular interaction, a Dual-Luciferase Reporter Assay was performed as the key validation step.
- Vector Construction: Researchers created two Firefly luciferase reporter vectors: one linked to the normal (Wild-Type, WT) 3' UTR of the AIFM3 gene, and a control vector where the predicted miR-210 binding site was mutated (MUT).
- Cell Preparation: Heart muscle cells (cardiomyocytes) were co-transfected with one of the Firefly reporters and a Renilla control plasmid.
- Treatment: The reporter cells were then treated with the therapeutic, miR-210-rich exosomes.
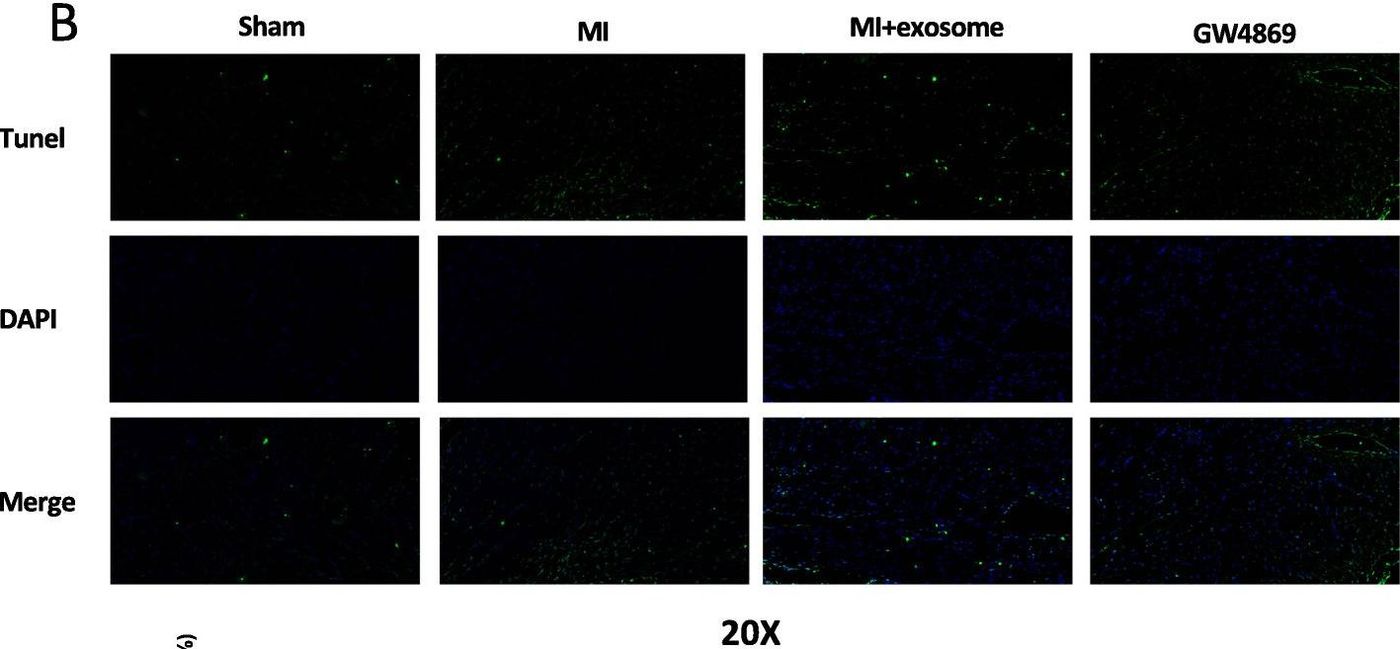 Figure 3. Immunohistochemistry showing reduced number of apoptotic cardiomyocytes in tissue samples treated with exosomes compared to MSCs-GW4869-exosomes or PBS. (Cheng H, et al., 2020)
Figure 3. Immunohistochemistry showing reduced number of apoptotic cardiomyocytes in tissue samples treated with exosomes compared to MSCs-GW4869-exosomes or PBS. (Cheng H, et al., 2020)
Key Findings:
- In cells containing the WT AIFM3 reporter, treatment with the miR-210-rich exosomes caused a significant decrease in normalized luciferase activity. This demonstrated that the exosome-delivered miR-210 was biologically active and successfully repressed its target gene.
- In cells containing the MUT AIFM3 reporter, the exosomes had no significant effect on luciferase activity. This crucial control proved that the interaction was specific to the predicted miR-210 binding site on the AIFM3 gene.
Conclusion: The dual-luciferase reporter assay provided the definitive molecular proof. It confirmed that the therapeutic, anti-apoptotic effect of the exosomes was mediated by the direct binding of their cargo, miR-210, to its functional target gene, AIFM3.
Ready to prove exosome cargo function? We design dual luciferase reporters with WT and MUT controls and deliver readouts confirming target engagement. Contact us for a free consultation.
References
- Sun Y, Li B, Song B, et al. CREB1/CRTC2 regulated tubular epithelial-derived exosomal miR-93-3p promotes kidney injury induced by calcium oxalate via activating M1 polarization and macrophage extracellular trap formation. J Nanobiotechnology. 2025 Mar 12;23(1):204.
- Cheng H, Chang S, Xu R, et al. Hypoxia-challenged MSC-derived exosomes deliver miR-210 to attenuate post-infarction cardiac apoptosis. Stem Cell Res Ther. 2020 Jun 8;11(1):224.
Frequently Asked Questions
For any inquiries, our support team is ready to help you get technical support for your research and maximize your experience with Creative Biostructure.