Exosomal RNA Microarray Services
Exosome microarray services leverage advanced chip technology to perform high-throughput, high-precision detection and quantitative analysis of diverse RNAs carried by exosome samples. This serves as a powerful tool for exploring disease mechanisms and discovering biomarkers. Creative Biostructure offers cutting-edge exosome RNA microarray services that enable precise analysis of RNA expression patterns, providing valuable insights for biomedical research and clinical applications.
What is Exosomal RNA Microarray and What are its Applications?
An exosomal RNA microarray is a molecular diagnostic technique used to profile and quantify various RNAs, such as mRNAs, miRNAs, and lncRNAs, that are contained within exosomes, which are a type of extracellular vesicle. Using pre-designed probes immobilized on a chip to bind specific RNA sequences, this method provides valuable insights into intercellular communication, disease mechanisms, and the identification of biomarkers for early detection of diseases including cancer.
The applications of exosomal microarrays include
- Biomarker Discovery
Exosomal RNAs act as diagnostic biomarkers, enabling disease detection, treatment response prediction, and identification of early recurrence.
- Cell-to-Cell Communication
Communication: Microarrays elucidate how RNA-containing exosomes transfer between cells to mediate intercellular communication and alter recipient cell functions.
- Diagnosis
Detecting exosomal RNA in non-invasive samples (e.g., serum) offers promise for early and personalized diagnostics in diseases like cancer.
- Research
Exosomal RNA microarrays enable high-throughput screening of thousands of RNA molecules simultaneously, essential for studying complex and scarce exosomal RNA populations.
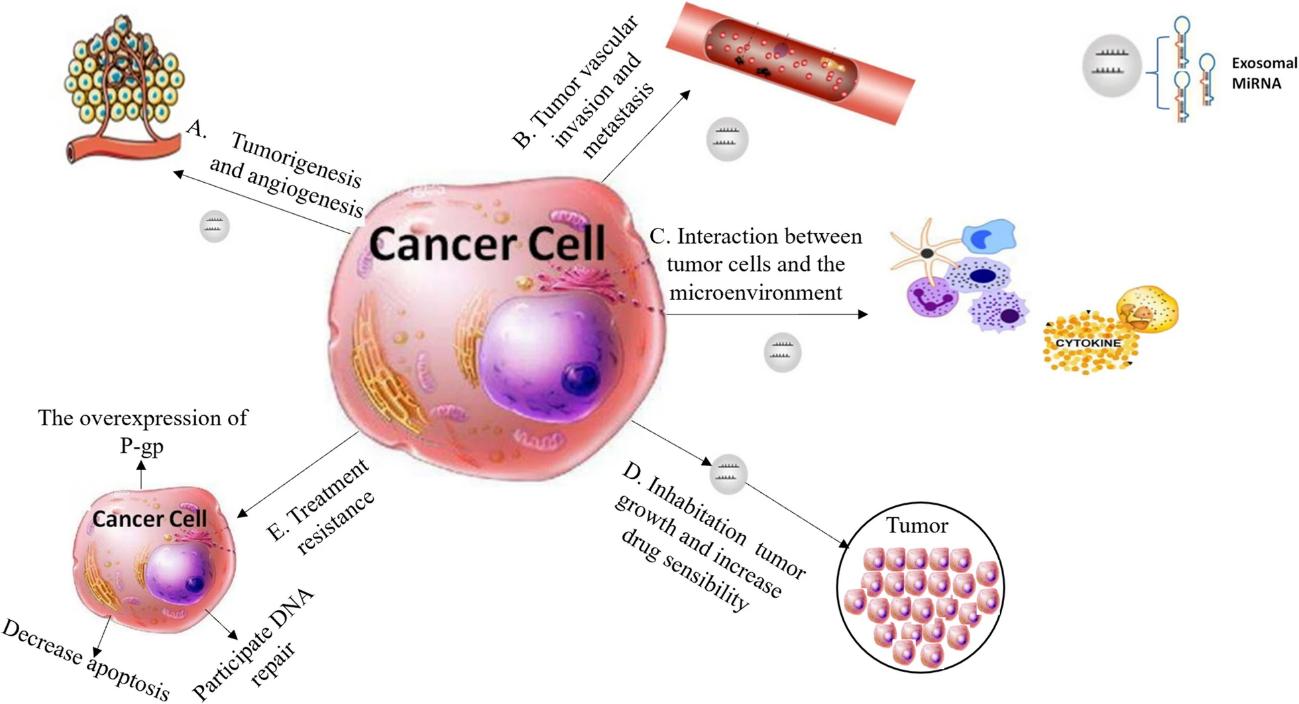
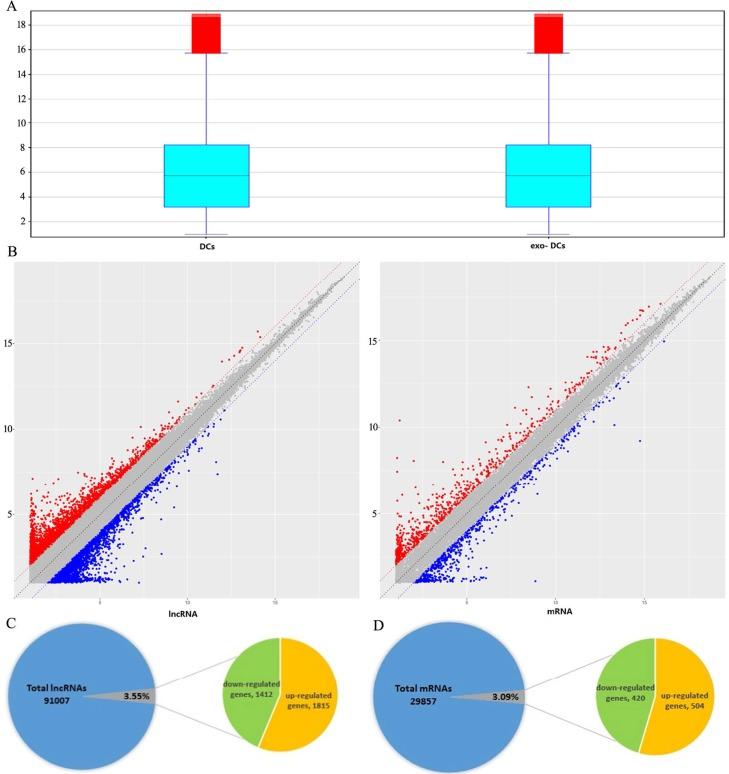 Figure 1. Summary of microarray results of lncRNA and mRNA expression in PEX-treated DCs (exo-DCs). (Chen J, et al., 2018)
Figure 1. Summary of microarray results of lncRNA and mRNA expression in PEX-treated DCs (exo-DCs). (Chen J, et al., 2018)
Our Exosomal RNA Microarray Services
Creative Biostructure provides an integrated exosomal RNA microarray service, delivering a complete workflow for precise transcriptomic analysis of exosome-derived RNA. Our process begins with optimized exosome isolation and RNA extraction protocols that ensure high sample purity and biological relevance. After stringent quality control, RNA is labeled and hybridized using advanced microarray platforms for high-resolution profiling. We also offer sophisticated bioinformatics support for in-depth interpretation, including expression analysis, functional classification, and interaction network mapping-enabling robust biomarker discovery and mechanistic insights in fields such as oncology, neuroscience, and regenerative medicine. Tailored for both exploratory and validation studies, our service is ideal for researchers leveraging exosomal RNA in biomarker development, drug response studies, or disease mechanism research.
Workflow of Exosomal RNA Microarray
Creative Biostructure ensures high sensitivity and reproducibility through standardized operating procedures and an industry-leading microarray platform.
Exosome Separation and Purification
Isolate and extract exosomes from biological samples such as serum and cell supernatant to ensure the structural integrity of exosomes and the representativeness of the samples.
RNA Extraction and Quality Control
High-quality total RNA is extracted from exosomes using established commercial kits and in-house optimized protocols, followed by rigorous quality control including assessments of integrity and purity.
RNA Labeling
RNA that meets quality standards is fluorescently labeled for subsequent microarray hybridization analysis.
Microarray Hybridization
Hybridize the marked exosomal RNA with probes on a customized microarray chip, using a high-throughput platform to capture the specific binding of target RNA molecules to the probes.
Detection and Analysis
Detect hybridization signals using high-resolution scanners to obtain raw RNA expression profile data, then perform expression quantification, differential analysis, and functional annotation through specialized bioinformatics workflows.
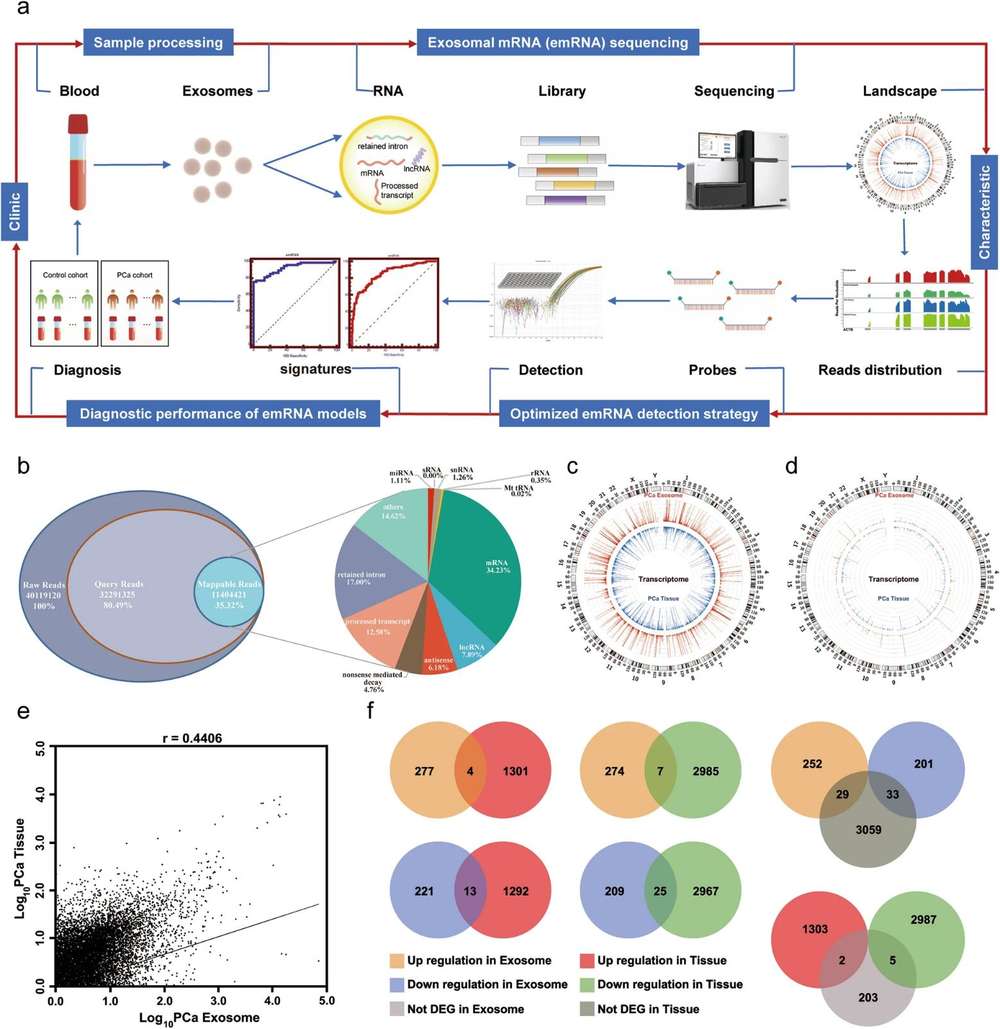
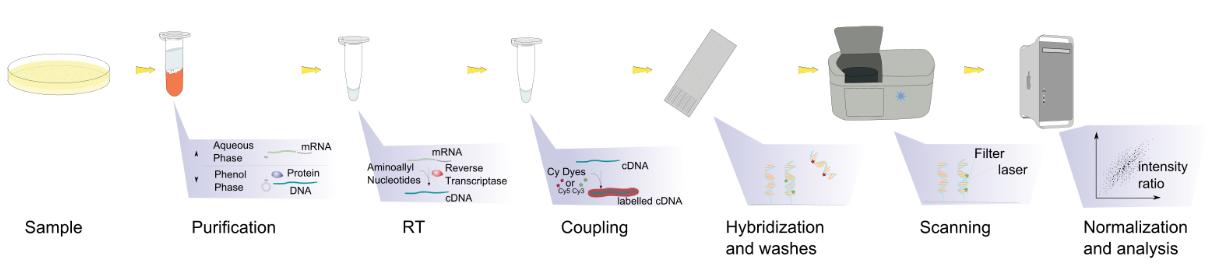 Figure. 2 Workflow of exosomal RNA microarray. (Creative Biostructure)
Figure. 2 Workflow of exosomal RNA microarray. (Creative Biostructure)
Supported Sample Range
To ensure your research obtains reliable and high-quality data results, Creative Biostructure provides comprehensive and flexible sample type support. The sample types we support include, but are not limited to:
| Sample Type | Minimum Requirement | Storage Conditions |
|---|---|---|
| Serum/Plasma | 200 μL | -80°C, avoid repeated freeze-thaw cycles |
| Cell Culture Supernatant | 5 mL | -80°C, requires pretreatment to remove cell debris |
| Urine | 10 mL | 4°C (within 24 hours)/-80°C (long-term storage) |
| Tissue Homogenate | 100 mg | -80°C, liquid nitrogen rapid freezing |
Final Deliverables
- Raw data files
- Standardized data report (including PCA, volcano plot, clustering heatmap)
- Differentially expressed RNA list
- Experimental protocol documentation (including exosome characterization, RNA QC report, compliant with ISEV guidelines)
- Data analysis manual (including R packages used and parameter settings)
Advantages of Our Exosomal RNA Microarray Services
- Standardized Processing of Exosomes and RNA
Stringent protocols for isolating exosomes and extracting RNA guarantee consistent sample quality and reliable data across all batches.
- Advanced Microarray Platforms
Our high-resolution, high-density microarray chips provide the capacity for extensive transcriptome coverage and dependable detection of expression variations.
- Integrated Bioinformatics Support
Facilitated by robust downstream analysis, which includes normalization, differential expression profiling, pathway enrichment, and data visualization, enabling deep biological interpretation.
- Customizable Solutions
Our services are customized to your specific research objectives, such as particular disease models, tissue types, or targeted transcript panels.
Case Study
Case: miRNA Profiling of Urinary Exosomes in a 5XFAD Mouse Model of Alzheimer's Disease
Background
To explore the potential noninvasive earlier biomarkers of AD in a 5XFAD mouse model, microRNAs (miRNAs) from urinary exosomes in 1-month-old pre-Aβ accumulation 5XFAD mice models and their littermate controls were profiled by microarray analysis. The differentially expressed miRNAs were further analyzed via droplet digital PCR (ddPCR).
Methods
- Preparation of urine samples
All urinary exosome samples were extracted from 30 mL of urine using the Exosome Isolation Q3 kit.
- Exosome isolation
Isolation of exosomes using the Q3 Exosome Isolation Kit.
- MicroRNA microarray
Total RNA was isolated using the miRNeasy Micro Kit. Each RNA sample was then used to generate biotinylated cRNA targets for the GeneChip miRNA 4.0 Array.
- Microarray data analysis
The raw data underwent global normalization (TAC software) due to the lack of reliable endogenous controls for exosomal miRNAs. Differential miRNA expression was identified (threshold: FC ≥1.2), and their target genes were predicted. Subsequent Gene Ontology (GO) and KEGG pathway enrichment analyses were performed to elucidate the biological functions and roles of these target genes, with a specific focus on pathways related to neurodegenerative and nervous system diseases.
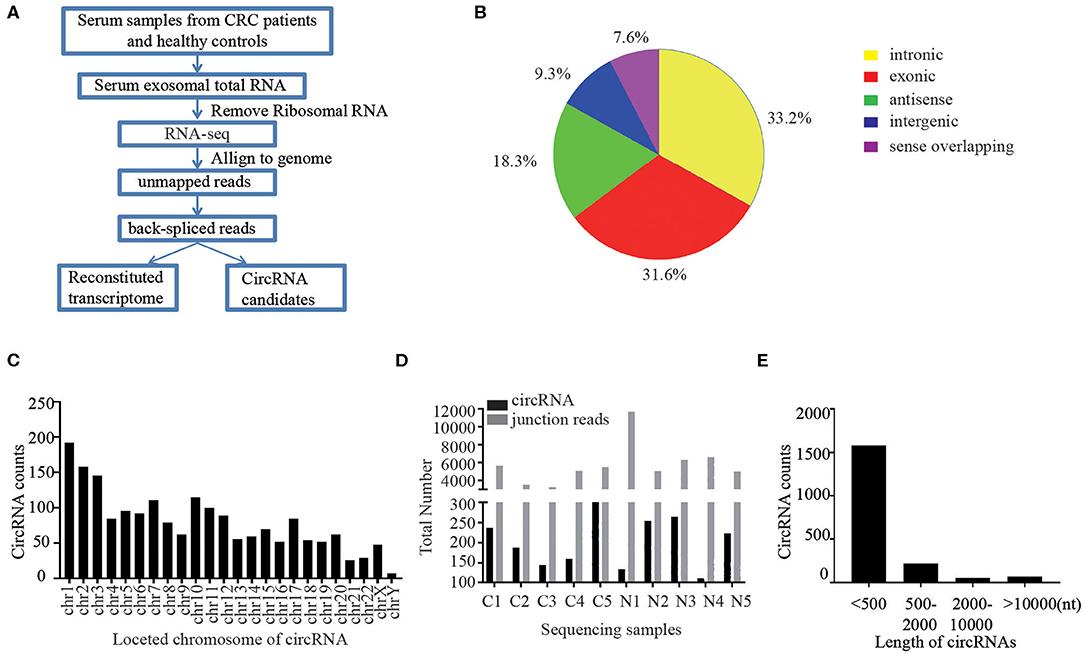
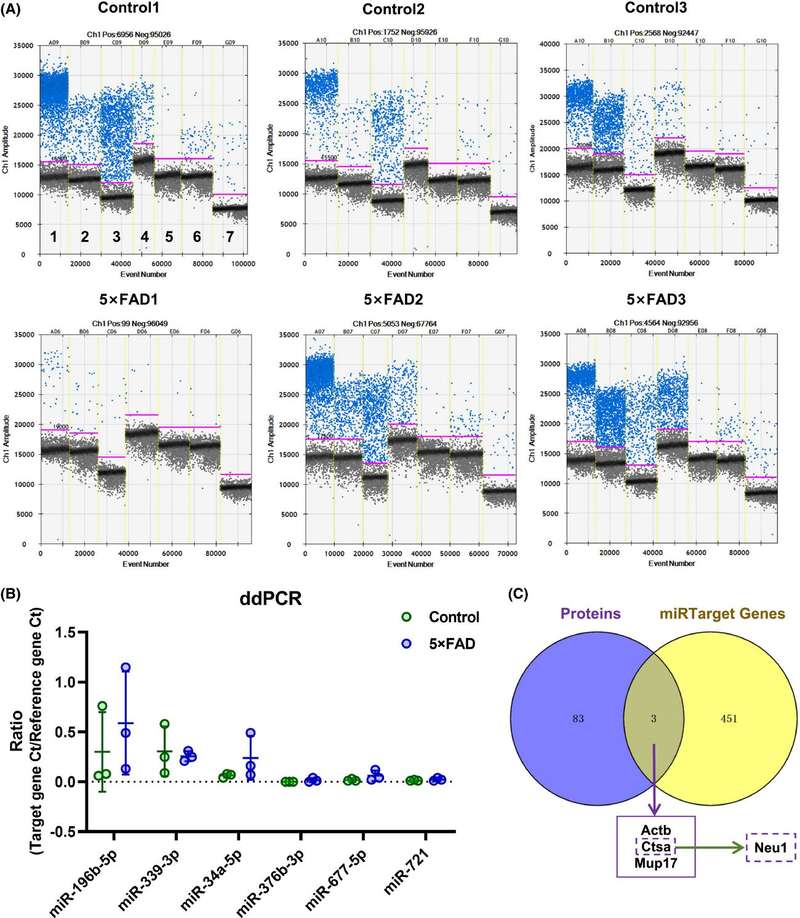 Figure. 3 Differentially expressed candidate miRNA in the urinary exosome miRNA microarray analysis of a 5XFAD mice model of AD. (Song Z, et al., 2021)
Figure. 3 Differentially expressed candidate miRNA in the urinary exosome miRNA microarray analysis of a 5XFAD mice model of AD. (Song Z, et al., 2021)
Conclusion
Microarray analysis identified 48 differentially expressed miRNAs (18 upregulated, 30 downregulated) in urinary exosomes before Aβ-plaque deposition. Six of these (miR-196b-5p, miR-339-3p, miR-34a-5p, miR-376b-3p, miR-677-5p, and miR-721) targeted genes and pathways linked to Alzheimer's disease, as validated by ddPCR. These miRNAs may serve as noninvasive biomarkers for early AD prevention in clinical settings.
Creative Biostructure leverages years of specialized expertise to deliver efficient, precise, and comprehensive exosome RNA microarray solutions. From exosome isolation, RNA extraction, and quality control to full transcriptome chip detection and in-depth bioinformatics analysis, we employ standardized protocols and stringent quality control measures to ensure data reliability and reproducibility. Contact us for further details, we are dedicated to serving your needs.
References
- Chen J, Wang S, Jia S, et al. Integrated analysis of long non-coding RNA and mRNA expression profile in pancreatic cancer derived exosomes treated dendritic cells by microarray analysis. Journal of Cancer. 2018, 9(1): 21.
- Song Z, Qu Y, Xu Y, et al. Microarray microRNA profiling of urinary exosomes in a 5XFAD mouse model of Alzheimer's disease. Animal models and experimental medicine. 2021, 4(3): 233-242.
Frequently Asked Questions
For any inquiries, our support team is ready to help you get technical support for your research and maximize your experience with Creative Biostructure.