What is Exosomal Proteomic Analysis?
In recent years, with the continuous development of science and technology, exosome proteomics assay, as an emerging technology, has attracted extensive attention in biopharmaceuticals. Exosomes are a kind of nanovesicles released by cells containing a variety of proteins, nucleic acids, and bioactive molecules. They can be transmitted to distant tissues and cells through body fluids such as blood, urine, and saliva, thus becoming a potential biomarker for disease diagnosis and treatment research.
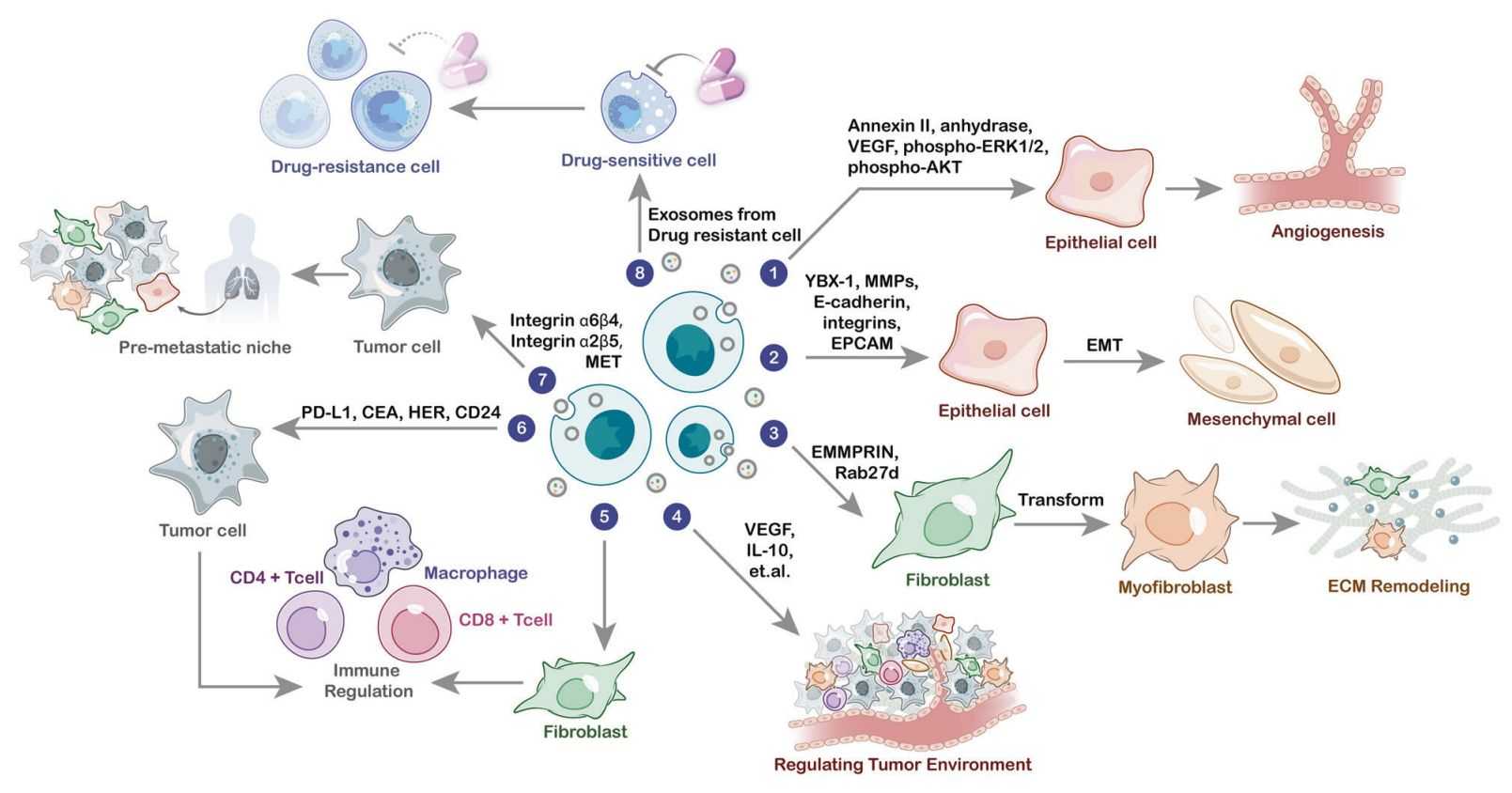 Figure 1. The role of exosomal proteins in cancer biology. (Wang X, et al., 2022)
Figure 1. The role of exosomal proteins in cancer biology. (Wang X, et al., 2022)
Principles of Exosome Proteomic Assays
Exosomes are a kind of nanovesicles released by cells, which are widely present in body fluids, such as plasma, urine, saliva, breast milk, etc., and can span various tissues and organs in organisms, playing an important role in intercellular signaling. Exosomes are rich in biologically active molecules, including proteins, nucleic acids, and lipids. The composition and content of these molecules change under different physiological or pathological conditions, making exosomes a potential biomarker.
The general workflow of an exosomal proteomics assay involves the following steps.
1. Exosome Isolation
Isolation of exosomes from biological samples such as cell culture media, blood, or other body fluids using ultracentrifugation, size exclusion chromatography, or immunoaffinity.
2. Protein Extraction and Digestion
The proteins within the isolated exosomes are extracted and then enzymatically digested, typically using proteases like trypsin, to generate peptides that can be analyzed by mass spectrometry.
3. Peptide Separation and Mass Spectrometry
The digested peptides are separated using techniques like liquid chromatography and then analyzed by mass spectrometry, which can identify and quantify the proteins based on the mass-to-charge ratio of the peptides.
4. Protein Identification and Quantification
The mass spectrometry data is analyzed using bioinformatics tools and databases to identify the proteins present in the exosome samples and determine their relative abundance.
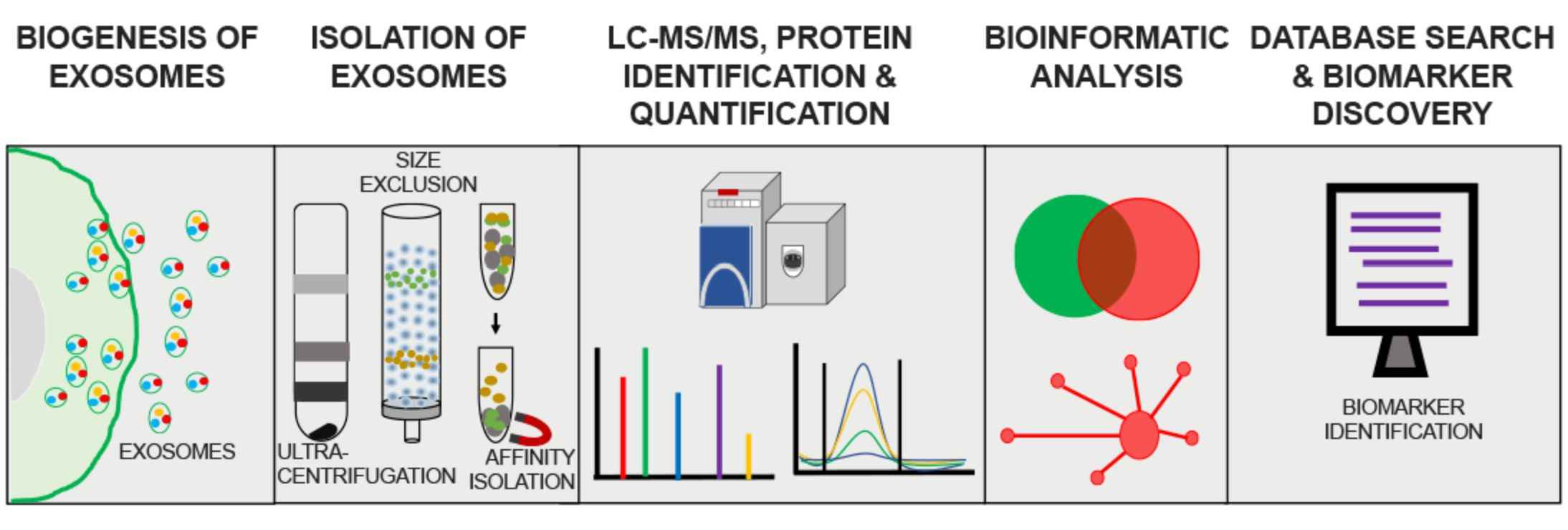 Figure 2. Flowchart of exosome proteomics assay. (Mathew B, et al., 2021)
Figure 2. Flowchart of exosome proteomics assay. (Mathew B, et al., 2021)
Commonly Used Techniques for Exosome Proteomics Testing
Peptide Separation Techniques
- Liquid Chromatography (LC) - Digested peptides are separated using High-Performance Liquid Chromatography (HPLC) or Ultra High-Performance Liquid Chromatography (UHPLC) before mass spectrometry analysis.
- Capillary Electrophoresis (CE) - Peptides can also be separated using capillary electrophoresis based on their charge-to-mass ratio.
Mass Spectrometry (MS)
- Tandem Mass Spectrometry (MS/MS) - The most commonly used technique in which peptides are ionized and then fragmented in a mass spectrometer to determine their amino acid sequence and identify the corresponding protein.
- Data-Dependent Acquisition (DDA) - In this method, the mass spectrometer automatically selects the most abundant peptide ions for fragmentation and identification.
- Data Independent Acquisition (DIA) - This technique analyzes all peptide ions in a sample regardless of their abundance, providing a more comprehensive and unbiased analysis of the exosome proteome.
Bioinformatics and Data Analysis
- Database Search - Acquired mass spectrometry data is matched against a protein sequence database to identify proteins present in exosomal samples.
- Non-Labeled Quantification - Determine the relative abundance of proteins based on the intensity of the peptide signal or the number of identified peptides.
- Isotope Labeling - Labeling of samples with isotope tags (e.g., TMT, iTRAQ) for multiplexed quantitative analysis of exosomal proteins under different conditions.
Exosome Proteomics Assays in Biopharmaceuticals
Biomarker Discovery and Validation
- Exosomes can be a source of potential biomarkers for various diseases, as their protein cargo reflects the state of the cell of origin.
- Exosome proteomics can be used to identify disease-specific protein signatures that can help in early detection, diagnosis, and monitoring of disease progression.
- These biomarkers can be further validated and developed as diagnostic or prognostic tools for biopharmaceutical applications.
Drug Target Identification and Validation
- Exosomal proteomics can provide insights into the molecular mechanisms of disease pathogenesis, thus helping to identify new drug targets.
- By analyzing changes in the exosome proteome after drug treatment, researchers can validate the targeting effect of therapeutic candidates and better understand their mechanism of action.
Exosome-based Drug Delivery
- Exosomes are being explored as natural nanocarriers for delivering therapeutic agents, such as small molecules, proteins, nucleic acids, etc.
- Exosome proteomics helps to characterize the targeting and loading properties of exosomes, leading to the development of more effective exosome-based drug delivery systems.
Research Case
- Proteomic Analysis of Exosomes Identifies Proteins Associated with Breast Cancer Metastasis
Proteomic analysis of cancer cell-derived exosomes can elucidate intercellular communication and identify candidate biomarkers for cancer diagnosis and treatment. Researchers conducted a comprehensive quantitative proteomic study of exosomes isolated from immortalized breast epithelial cells and tumor cell lines. A total of 2135 unique proteins and several metastasis-specific markers are quantified with high confidence. These metastasis-specific markers are highly correlated with survival in breast cancer (BC) patients. These data have contributed to the elucidation of the molecular mechanisms underlying primary tumor development and progression.
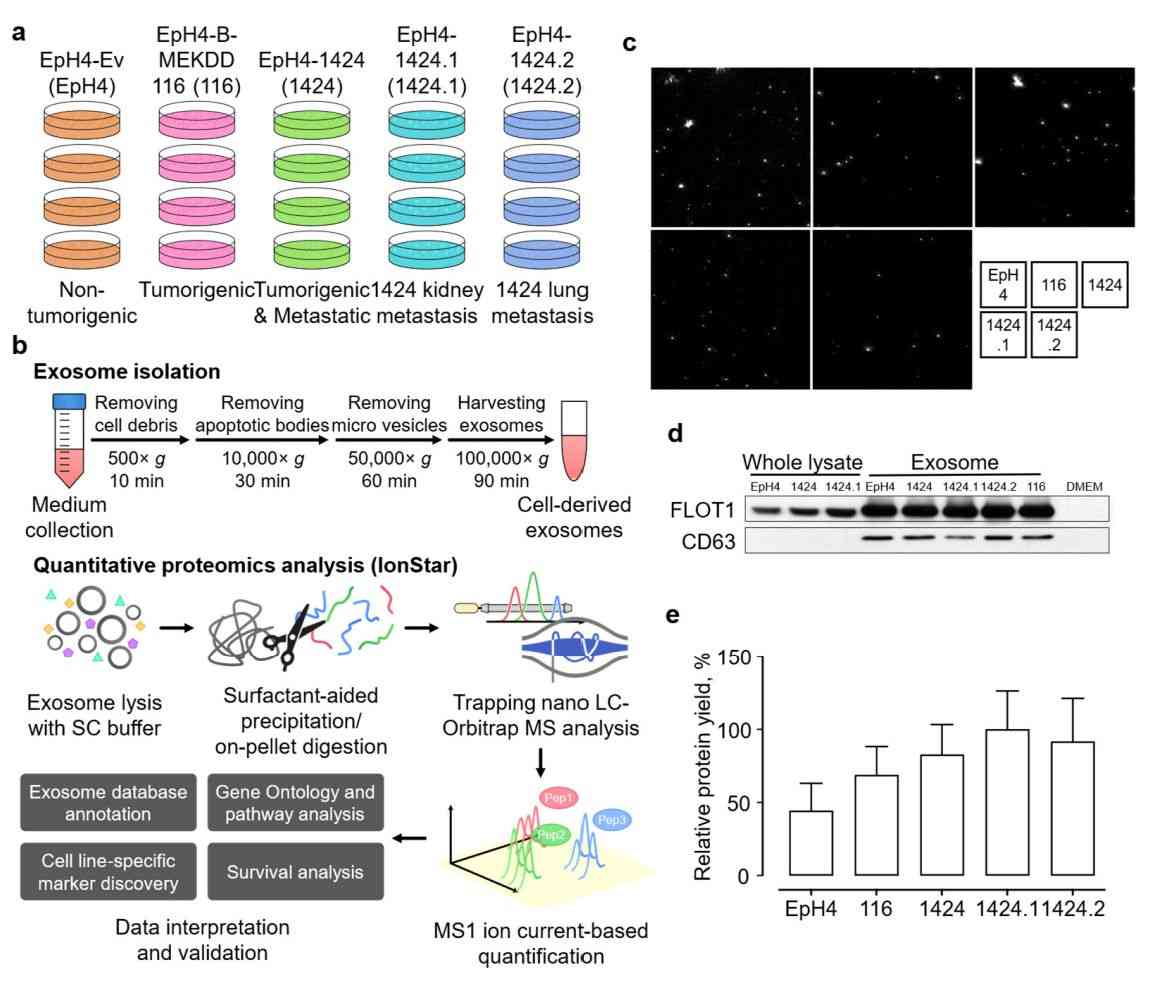 Figure 3. Quantitative proteomics of exosomes isolated from the EpH4-Ev cell line. (Shen S, et al., 2023)
Figure 3. Quantitative proteomics of exosomes isolated from the EpH4-Ev cell line. (Shen S, et al., 2023)
- Proteomic Analysis of Human Mesenchymal Stem Cell Exosomes
In recent years, mesenchymal stem cell (MSC)-derived exosomes have shown a wide range of applications. Researchers separated MSCs-Exos from different tissue sources by ultracentrifugation and performed proteomic analysis, which detected 1014 proteins. Bioinformatics analysis showed that bone marrow MSC-derived exosomes have strong regenerative ability, adipose tissue MSC-derived exosomes are involved in immunomodulation, and umbilical cord MSC-derived exosomes play a role in tissue injury repair.
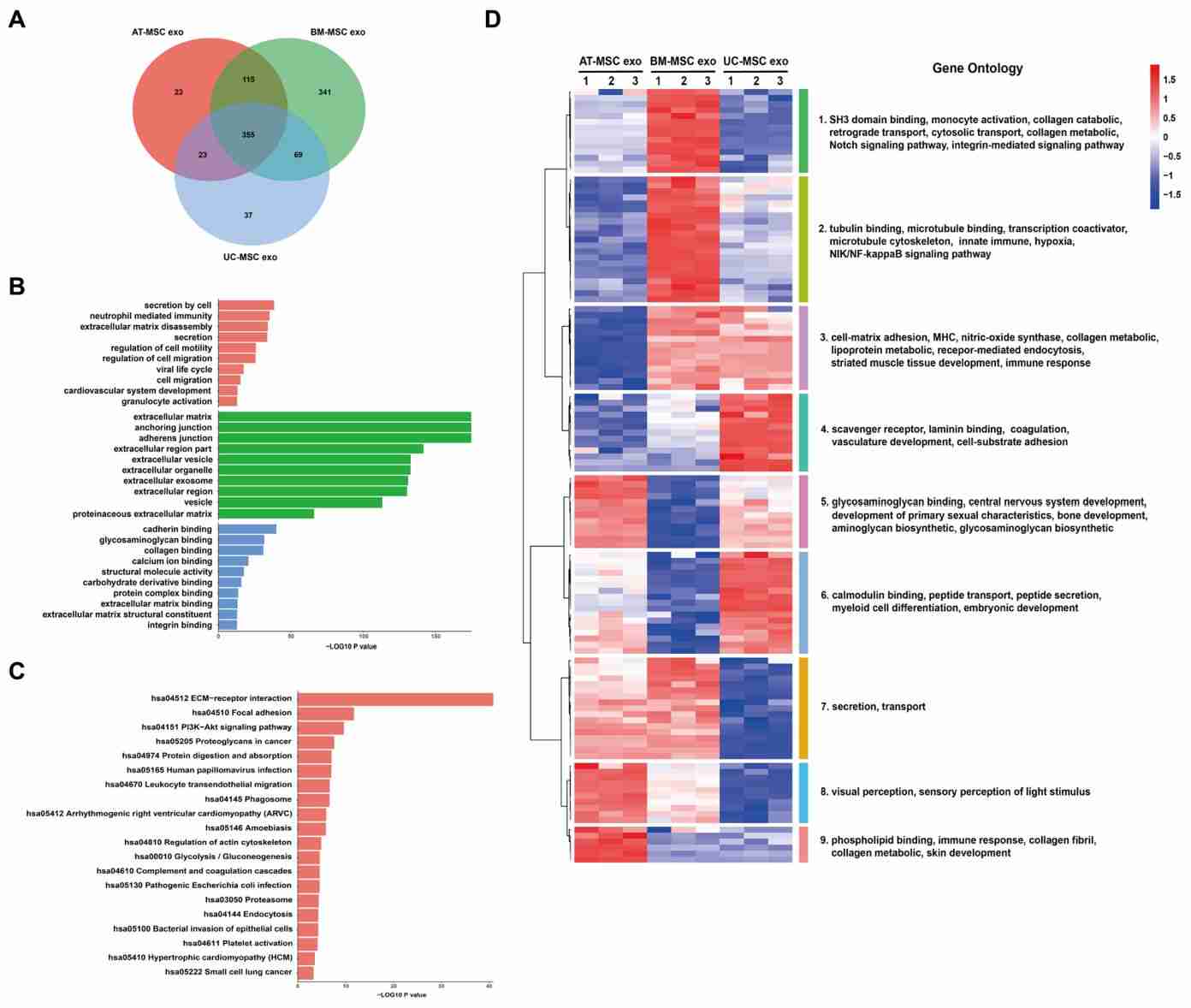 Figure 4. Bioinformatics analysis of exosomal proteins from three mesenchymal stem cells. (Wang ZG, et al., 2020)
Figure 4. Bioinformatics analysis of exosomal proteins from three mesenchymal stem cells. (Wang ZG, et al., 2020)
Our Services and Products
The development of exosomal proteomic assays has created tremendous opportunities and challenges for the biopharmaceutical field. By gaining a deeper understanding of the composition and function of proteins in exosomes, Creative Biostructure can better assist clients in developing new drugs and diagnostic tools. If you have any questions about our services, please feel free to contact us.
References
- Wang X, et al. The updated role of exosomal proteins in the diagnosis, prognosis, and treatment of cancer. Exp Mol Med. 2022. 54(9): 1390-1400.
- Mathew B, et al. Exosomes as Emerging Biomarker Tools in Neurodegenerative and Neuropsychiatric Disorders—A Proteomics Perspective. Brain Sciences. 2021. 11(2): 258.
- Shen S, et al. Comparative Proteomics Analysis of Exosomes Identifies Key Pathways and Protein Markers Related to Breast Cancer Metastasis. International Journal of Molecular Sciences. 2023. 24(4): 4033.
- Wang ZG, et al. Comprehensive proteomic analysis of exosomes derived from human bone marrow, adipose tissue, and umbilical cord mesenchymal stem cells. Stem Cell Res Ther. 2020. 11(1): 511.