Membrane Scaffold Proteins (MSPs) for Mempro™ Nanodisc
Creative Biostructure has established a comprehensive nanodisc platform based on our Mempro™ Membrane Protein platform during the past decades. Mempro™ Nanodisc technique shows several advantages in membrane protein production, membrane protein structural investigation and functional characterization over the traditional methods. One of our magic nanodisc products is various membrane scaffold proteins (MSPs) including Apo-A1, MSP1D1, and MSP1E3D1. Furthermore, assembling nanolipoproteins with APO-E4 has also been possible to incorporate membrane proteins such as bacteriorhodopsin (bR). There also exists other naturally occurring proteins forming amphipathic helices that potentially could be used to make other MSP constructs such as: synucleins, apolipophorins, melittin and apomyoglobin.
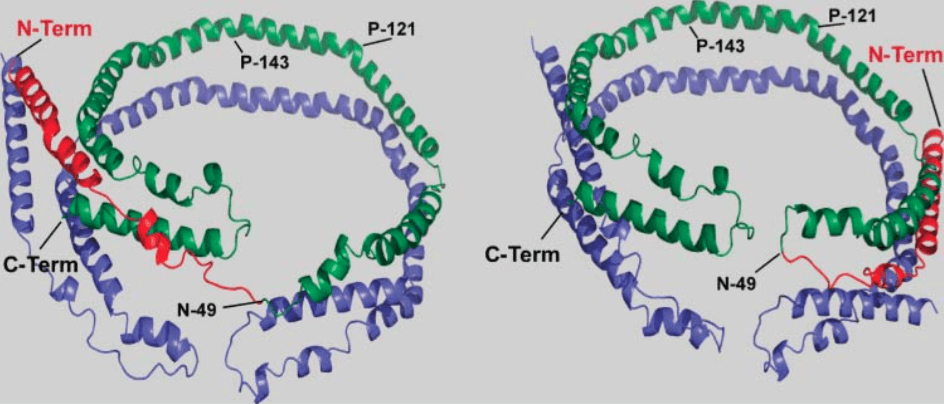 Figure 1. Two 3D models for apoA-I molecules with a flexible N-terminal end and a “belt-buckle” conformation (Journal of Lipid Research, 2008).
Figure 1. Two 3D models for apoA-I molecules with a flexible N-terminal end and a “belt-buckle” conformation (Journal of Lipid Research, 2008).
MSPs are typically membrane proteins which are applied in the reconstitution of purified proteins. The most famous and used membrane scaffold protein is the apo-A1.
The 243 amino acid human apo-A1 is composed of a N-terminal globular domain followed by 10 x 22-mer amphipathic helical repeats, 8 of which contain 20 amino acid residues and 2 containing 10 residues. The globular domain consists of four sub domains denoted G0 to G3. The most interesting feature of the apo-A1 protein related to phospholipid nanodiscs is the ability of the lipid associated domain to stabilize phospholipids in solution. Apo-A1 is water soluble but upon binding the protein undergoes a significant structural transition changing from around 50% alpha helix, in the lipid free state, to around 75% upon binding.
Creative Biostructure can provide a variety of purified MSPs (with or without a his-tag) which are able to assemble into custom nanodiscs with varying sizes. You can easily detect the his-tagged MSPs of the nanodiscs for characterization, but if your targeted protein which contains a his-tag itself, untagged MSPs are recommended. Mouse homologue membrane scaffold proteins are also available because human MSPs might interfere with certain cases sometimes such as immunization of mice with human antigens.
Creative Biostructure offers a variety of membrane scaffold proteins which are lyophilized and stable to combine with various phospholipids. Mempro™ Nanodisc platform will also provides the expert support and protocols in details. Membrane Scaffold Proteins (MSPs) for Mempro™ Nanodisc are as belows:
- MSP1D1 lyophilized protein with a his-tag (1mg/5mg)
- MSP1D1 dH5 lyophilized protein with a his-tag (1mg/5mg)
- MSP1E3D1 lyophilized protein with a his-tag (1mg/5mg)
- MSP2N2 lyophilized protein with a his-tag (1mg/5mg)
- Mouse MSP1D1 lyophilized protein with a his-tag (1mg/5mg)
- Mouse MSP1E3D1 lyophilized protein with a his-tag (1mg/5mg)
Mempro™ Nanodisc Technology is an invaluable tool to study the structure and function of membrane proteins, and there are a lot of applications based on Mempro™ Nanodisc Technology. A variety of Mempro™ Nanodisc products are also availble, please see the related sections below for more information and feel free to contact us for a detailed quote.
References:
I. G. Denisov and S. G. Sligar. (2016). Nanodiscs for structural and functional studies of membrane proteins. Nature Structural & Molecular Biology,23: 481–486.
M. Thomas, et al. (2008). Three-dimensional models of HDL apoA-I: implications for its assembly and function. Journal of Lipid Research, 49(9):1875– 1883.
A. Y. Shih, et al. (2005). Molecular dynamics simulations of discoidal bilayers assembled from truncated human lipoproteins. Biophysical Journal, 88(1):548–556.