Structural Research of Dicistroviridae
The family of Dicistroviridae is a group of small, non-enveloped viruses containing three genera, Cripavirus, Triatovirus, and Aparavirus, with two open reading frames (ORFs) or cis-retroviruses in their positive single-stranded RNA (+ssRNA) genomes. All viruses in the family infect arthropod hosts, some of which have devastating economic consequences, such as the Kashmir bee virus (KBV) and mud crab dicistrovirus (MCDV). High-resolution structures of pathogenic viruses are essential for molecular understanding and the development of new methods of disease prevention. In recent years, researchers have reported the three-dimensional (3D) structures of several members of the Dicistroviridae by cryo-electron microscopy (cryo-EM), contributing to the development of antiviral inhibitors.
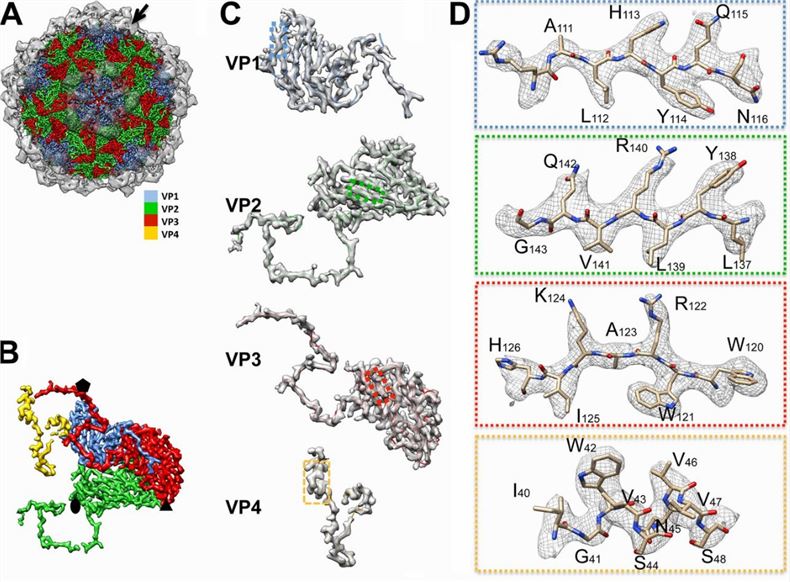 Figure 1. Structural features of mud crab dicistrovirus (MCDV). (Gao Y, et al., 2019)
Figure 1. Structural features of mud crab dicistrovirus (MCDV). (Gao Y, et al., 2019)
Overview of Dicistroviridae Virus Particles
Dicistroviridae virus particles are roughly spherical, unenveloped, and have a particle size of approximately 30 nm. icosahedra that exhibit pseudo-T=3 symmetry consist of 60 protomers, each containing a single molecule of VP1, VP2, and VP3. vP4 is a peptide of 70 to 90 residues in length that is attached to the inner surface of the parvoviral and short-viral capsids and is in contact with the genome. 5 ' ORFs encode nonstructural proteins, including viral polymerases, proteases, and deconjugates, whereas 3' ORFs encode multiprotein precursors of the mature capsid proteins VP1, VP2, VP3, and VP4.
Structural Features of the Constituent Proteins of Dicistroviridae
The researchers demonstrated the structural features of the constituent proteins of Dicistroviridae by 3.5 Å mapping of MCDV. The structural proteins VP1, VP2, and VP3 share a "jelly roll" fold with the β chain. The VP1 forms a pentamer around 5-fold axes, and VP2 and VP3 form alternating heterohexamers around icosahedral 3-fold axes. The C-terminus and CD-loop of each VP1 are located away from the surface of the capsid, while the N-terminus is inserted into the capsid's inner surface. The N-terminus of VP2 connects the neighboring VP2 and VP3 subunits. The N-terminal region of VP3 twists around the 5-fold axes and interacts to form a cylindrical structure. This is a common feature of Dicistroviridae members.
Creative Biostructure offers highly purified virus-like particles (VLPs) products for use in viral structure research. Our full range of products helps clients to understand the three-dimensional structure of the viral capsid and how it interacts with nucleic acids, which is of great application value in unraveling the mechanism of the viral life cycle, analyzing exogenous-mediated molecular delivery, antiviral drug, and vaccine development.
Knowledge of viral structure at atomic resolution is useful and powerful for the discovery and design of vaccines and antiviral drugs. By providing structural analysis services and a comprehensive range of virus-like particles (VLPs) products, Creative Biostructure furthers clients' scientific research into the structural biology of viruses. Our products contribute to a deeper understanding of viral biology, facilitate the discovery of antiviral drugs and vaccines, and support diagnostic advances.
| Cat No. | Product Name | Virus Name | Source | Composition |
| CBS-V572 | Drosophila VLP (copia Gag Proteins) | Yeast recombinant | Copia Gag | |
| Explore All Dicistroviridae Virus-like Particle Products | ||||
With our electron microscopy (EM) platform, Creative Biostructure can offer structural analysis services for virus particles and virus-like particles. Our scientists have the expertise and the necessary skills to unravel the complex structure of viruses and help clients understand the biology, replication mechanisms, and interaction patterns of viruses.
Whether it is structural analysis for a specific virus of interest or customized VLP construction, we are committed to providing a personalized service that meets the objectives and research priorities of each client. If you are interested in our services, please feel free to contact us. We look forward to working with you.
References
- Gao Y, et al. Cryo-electron Microscopy Structures of Novel Viruses from Mud Crab Scylla paramamosain with Multiple Infections. J Virol. 2019. 93(7): e02255-18.
- Mukhamedova L, et al. Virion structure and in vitro genome release mechanism of dicistrovirus Kashmir bee virus. J Virol. 2021. 95(11): e01950-20.
- Valles SM, et al. ICTV Virus Taxonomy Profile: Dicistroviridae. J Gen Virol. 2017. 98(3): 355-356.