Structural Research of Apical Sodium-Dependent Bile Acid Transporters
The apical sodium-dependent bile acid transporters (ASBTs) are integral membrane proteins that play a critical role in the uptake of bile acids from the small intestine. Recent structural studies have shed light on the molecular mechanisms of ASBT function and provided insights into its potential as a therapeutic target for bile acid-related disorders.
The crystal structures of ASBT from different species, including human, have been determined at high resolution, revealing the overall architecture of the protein and the location of the substrate-binding site. These studies have also identified key residues that are involved in substrate recognition and binding, as well as the sodium ion binding site. In addition, cryo-electron microscopy studies have provided insight into the conformational changes that occur during the transport cycle of ASBT. These studies have revealed a rocker-switch mechanism that is essential for the translocation of bile acids across the membrane. Furthermore, the use of pharmacological agents has helped to elucidate the binding mechanism and substrate specificity of ASBT. These studies have shown that ASBT can be targeted by small molecules to regulate bile acid transport and have potential for the development of novel therapeutics for bile acid-related disorders.
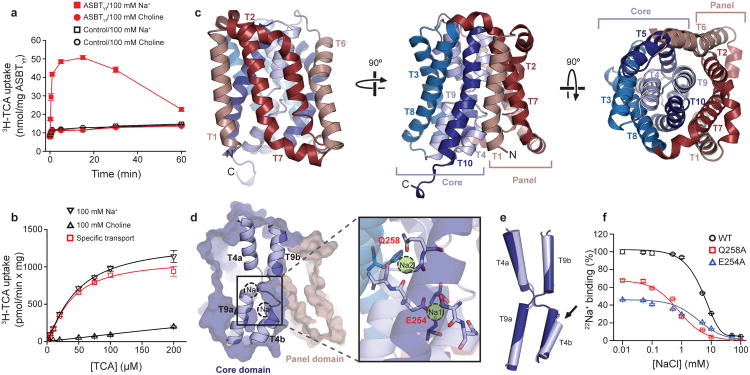 Figure 1. Crystal structure of ASBTYf. (Zhou X, et al., 2014)
Figure 1. Crystal structure of ASBTYf. (Zhou X, et al., 2014)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Bacterial homologue of ASBT (ASBTNM) with bound taurocholate (expressed in E. coli) | Neisseria meningitidis | X-ray diffraction | 2.20 Å | 3ZUY |
| Bacterial homologue of ASBT (ASBTYf), inward-open conformation (expressed in E. coli) | Yersinia frederiksenii | X-ray diffraction | 1.95 Å | 4N7W |
| Bacterial homologue of ASBT (ASBTγf), P10C-S291C mutant, inward-facing state complexed with citrate (expressed in E. coli) | Yersinia frederiksenii | X-ray diffraction | 1.85 Å | 6LGV |
| Bacterial homologue of ASBT (ASBTYf), cysteine-pair mutant (Y113C-P190C), outward facing state (expressed in E. coli) | Yersinia frederiksenii | X-ray diffraction | 2.86 Å | 6LH1 |
| Sodium taurocholate co-transporting polypeptide (NTCP) transporter (expressed in Spodoptera frugiperda) | Homo sapiens | Cryo-EM single particle analysis | 3.30 Å | 7FCI |
| NTCP transporter in complex with nanobody 87 (expressed in HEK293 cells) | Homo sapiens | Cryo-EM single particle analysis | 3.70 Å | 7PQG |
Table 1. Structural Research of Apical Sodium-Dependent Bile Acid Transporters (ASBT).
At Creative Biostructure, our services include a wide range of techniques such as X-ray crystallography, cryo-electron microscopy, and NMR spectroscopy. Our scientists have expertise in protein production, purification, and crystallization, as well as data collection, processing, and structure determination. We are committed to delivering timely and accurate results and providing excellent communication throughout the project.
We offer flexible and customized solutions to meet your needs, including full-service support, specific project-based work, and technology transfer. Our goal is to provide our clients with reliable and high-quality results, and to develop a long-term partnership to support their structural biology research needs. If you are interested in our services, please feel free to contact us, and we will offer professional and comprehensive solutions.
References
- Hu N J, et al. Crystal structure of a bacterial homologue of the bile acid sodium symporter ASBT. Nature. 2011, 478(7369): 408-411.
- Zhou X, et al. Structural basis of the alternating-access mechanism in a bile acid transporter. Nature. 2014, 505(7484): 569-573.
- Wang X, et al. Substrate binding in the bile acid transporter ASBTYf from Yersinia frederiksenii. Acta Crystallographica Section D: Structural Biology. 2021, 77(1): 117-125.
- Wang X, et al. An engineered disulfide bridge traps and validates an outward-facing conformation in a bile acid transporter. Acta Crystallographica Section D: Structural Biology. 2021, 77(1): 108-116.
- Park J H, et al. Structural insights into the HBV receptor and bile acid transporter NTCP. Nature. 2022, 606(7916): 1027-1031.
- Goutam K, et al. Structural basis of sodium-dependent bile salt uptake into the liver. Nature. 2022, 606(7916): 1015-1020.