Structural Research of Alpha-helical Pore-forming Toxins
Pore-forming toxins (PFTs) are important components of the molecular offensive and defensive machinery in many species. In eukaryotes, PFTs are primarily involved in the innate immune response, whilst in prokaryotes, they constitute the major virulence factor group of many pathogenic bacteria. By puncturing holes in the membrane, pathogenic PFTs can disrupt the osmotic balance of host target cells or insert secondary intracellular toxins through the pores formed in the membrane. Many pathogenic bacteria, including strains that are highly resistant to antibiotics, use PFTs in their invasive arsenal. This makes them attractive targets for developing therapies to reduce the acquired resistance that occurs with conventional antimicrobial therapy.
PFTs are produced in a soluble, often monomeric form and recognize target cells by binding to specific receptors, thereby concentrating the protein to the membrane surface prior to exposure of transmembrane hydrophobic regions, oligomerization, and membrane insertion. Depending on the secondary structure of the membrane assembly, PFTs can be classified into α-PFTs, which use amphipathic helical loops to construct the pore, or β-PFTs, in which the β-barrel is employed to traverse the membrane.
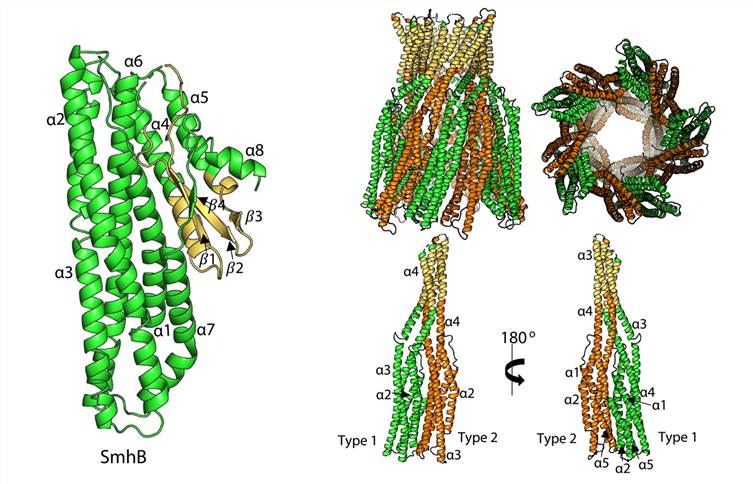 Figure 1. Structure of soluble and pore form SmhB. (Churchill-Angus A M, et al., 2021)
Figure 1. Structure of soluble and pore form SmhB. (Churchill-Angus A M, et al., 2021)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Cytolysin A pore (expressed in Escherichia coli) | Escherichia coli K-12 | X-ray diffraction | 3.29 Å | 2WCD |
| AhlB pore of the tripartite alpha-pore forming toxin (expressed in Escherichia coli) | Aeromonas hydrophila | X-ray diffraction | 2.94 Å | 6GRJ |
| Pre-pore AhlB (expressed in Escherichia coli) | Aeromonas hydrophila | X-ray diffraction | 2.55 Å | 6H2F |
| Soluble AhlB (expressed in Escherichia coli) | Aeromonas hydrophila | X-ray diffraction | 2.33 Å | 6GRK |
| Soluble AhlC form 1 (expressed in Escherichia coli) | Aeromonas hydrophila | X-ray diffraction | 2.35 Å | 6H2E |
| Soluble AhlC form 2 (expressed in Escherichia coli) | Aeromonas hydrophila | X-ray diffraction | 2.62 Å | 6H2D |
| Soluble AhlC triple head mutant (expressed in Escherichia coli) | Aeromonas hydrophila | X-ray diffraction | 1.92 Å | 6R1J |
| SmhB pore of the tripartite alpha-pore forming toxin (expressed in Escherichia coli) | Serratia marcescens | X-ray diffraction | 6.98 Å | 7A0G |
| Soluble SmhA crystal form 1 (expressed in Escherichia coli) | Serratia marcescens | X-ray diffraction | 2.98 Å | 7A26 |
| Soluble SmhA crystal form 2 (expressed in Escherichia coli) | Serratia marcescens | X-ray diffraction | 2.57 Å | 7A27 |
| Soluble SmhB (expressed in Escherichia coli) | Serratia marcescens | X-ray diffraction | 1.84 Å | 6ZZ5 |
| Soluble SmhB crystal form 2 (expressed in Escherichia coli) | Serratia marcescens | X-ray diffraction | 1.86 Å | 6ZZH |
| Full-length Cry6Aa (expressed in Pseudomonas fluorescens) | Bacillus thuringiensis | X-ray diffraction | 2.70 Å | 5KUD |
| Trypsin activated Cry6Aa (expressed in Pseudomonas fluorescens) | Bacillus thuringiensis | X-ray diffraction | 2.00 Å | 5KUC |
| FraC eukaryotic pore-forming toxin from sea anemone | Actinia fragacea | X-ray diffraction | 1.80 Å | 3LIM |
| FraC with lipids (expressed in Escherichia coli) | Actinia fragacea | X-ray diffraction | 3.14 Å | 4TSY |
| Soluble FraC monomer (I) (expressed in Escherichia coli) | Actinia fragacea | X-ray diffraction | 1.70 Å | 3VWI |
| Soluble FraC monomer (II) (expressed in Escherichia coli) | Actinia fragacea | X-ray diffraction | 2.10 Å | 3W9P |
| FraC dimer with phosphorylcholine (I) (expressed in Escherichia coli) | Actinia fragacea | X-ray diffraction | 1.60 Å | 4TSL |
| FraC dimer with phosphorylcholine (II) (expressed in Escherichia coli) | Actinia fragacea | X-ray diffraction | 1.57 Å | 4TSN |
| FraC with DHPC bound (I) (expressed in Escherichia coli) | Actinia fragacea | X-ray diffraction | 2.30 Å | 4TSO |
| FraC with DHPC bound (II) (expressed in Escherichia coli) | Actinia fragacea | X-ray diffraction | 2.15 Å | 4TSP |
| FraC with DHPC bound (III) (expressed in Escherichia coli) | Actinia fragacea | X-ray diffraction | 1.60 Å | 4TSQ |
| Hexameric anti-microbial peptide channel dermcidin | Homo sapiens | X-ray diffraction | 2.49 Å | 2YMK |
| TcdA1 (expressed in Escherichia coli) | Photorhabdus luminescens | X-ray diffraction | 3.50 Å | 4O9Y |
| TcdB2-TccC3 (expressed in Escherichia coli) | Photorhabdus luminescens | X-ray diffraction | 2.17 Å | 4O9X |
| Tc toxin TcdA1 in its pore state (obtained by flexible fitting) (expressed in Escherichia coli) | Photorhabdus luminescens | Cryo-EM single particle analysis | 3.46 Å | 5LKH |
| Tc toxin TcdA1 in its pore state (expressed in Escherichia coli) | Photorhabdus luminescens | Cryo-EM single particle analysis | 3.46 Å | 5LKI |
| Tc holotoxin pore (expressed in Escherichia coli) | Photorhabdus luminescens | Cryo-EM single particle analysis | 3.40 Å | 6SUF |
| Tc holotoxin pore, TccC3-D651A mutant (expressed in Escherichia coli) | Photorhabdus luminescens | Cryo-EM single particle analysis | 3.40 Å | 6SUE |
| Listeriolysin O pore-forming toxin | Listeria monocytogenes | X-ray diffraction | 2.15 Å | 4CDB |
| XaxAB pore complex (expressed in Escherichia coli) | Xenorhabdus nematophila | Cryo-EM single particle analysis | 4.00 Å | 6GY6 |
| XaxA monomer (expressed in Escherichia coli) | Xenorhabdus nematophila | X-ray diffraction | 2.50 Å | 6GY8 |
| XaxB monomer (expressed in Escherichia coli) | Xenorhabdus nematophila | X-ray diffraction | 3.40 Å | 6GY7 |
| VacA vacuolating cytotoxin A oligomeric assembly OA-1 | Helicobacter pylori | Cryo-EM single particle analysis | 3.20 Å | 6NYF |
| VacA oligomeric assembly OA-2a | Helicobacter pylori | Cryo-EM single particle analysis | 3.90 Å | 6NYG |
| VacA oligomeric assembly OA-2b | Helicobacter pylori | Cryo-EM single particle analysis | 3.20 Å | 6NYJ |
| VacA oligomeric assembly OA-2c | Helicobacter pylori | Cryo-EM single particle analysis | 3.70 Å | 6NYL |
| VacA oligomeric assembly OA-2d | Helicobacter pylori | Cryo-EM single particle analysis | 3.60 Å | 6NYM |
| VacA oligomeric assembly OA-2e | Helicobacter pylori | Cryo-EM single particle analysis | 3.50 Å | 6NYN |
| VacA hexamer | Helicobacter pylori | Cryo-EM single particle analysis | 3.80 Å | 6ODY |
| RhopH complex in soluble form | Plasmodium falciparum | Cryo-EM single particle analysis | 2.92 Å | 7KIY |
| Vip3Bc1 tetramer (expressed in Pseudomonas fluorescens) | Bacillus thuringiensis | Cryo-EM single particle analysis | 3.90 Å | 6YRF |
| Vip3Bc1 tetramer in processed, activated state (expressed in Pseudomonas fluorescens) | Bacillus thuringiensis | Cryo-EM single particle analysis | 4.80 Å | 6YRG |
| Vip3Bc1 tetramer in processed, activated state (expressed in Pseudomonas fluorescens) | Bacillus thuringiensis | Cryo-EM single particle analysis | 4.75 Å | 7NTX |
Table 1. Structural Research of Alpha-helical Pore-forming Toxins.
Due to the rapid advancement of cryo-electron microscopy (Cryo-EM), it has become a valuable tool for structural biology, especially in the pharmaceutical industry. Our Cryo-EM services allow you to analyze biomolecules accurately, including but not limited to ribosomes, subcellular organelles, extracellular vesicles, nucleic acids, cytoskeleton, membrane proteins, protein complexes, and antigen-antibody complexes.
Supported by years of experience, Creative Biostructure can also offer X-ray crystallography services in our state-of-the-art facilities and has developed an X-ray crystallography pipeline covering all technical stages, from gene synthesis to structure determination. If you are interested in our services, please feel free to contact our scientists, and they will offer professional and comprehensive solutions.
References
- Mueller M, et al. The structure of a cytolytic α-helical toxin pore reveals its assembly mechanism. Nature. 2009, 459(7247): 726-730.
- Wilson J S, et al. Identification and structural analysis of the tripartite α-pore forming toxin of Aeromonas hydrophila. Nature Communications. 2019, 10(1): 2900.
- Churchill-Angus A M, et al. Characterisation of a tripartite α-pore forming toxin from Serratia marcescens. Scientific Reports. 2021, 11(1): 1-15.
- Dementiev A, et al. The pesticidal Cry6Aa toxin from Bacillus thuringiensis is structurally similar to HlyE-family alpha pore-forming toxins. BMC Biology. 2016, 14(1): 1-16.
- Mechaly A E, et al. Structural insights into the oligomerization and architecture of eukaryotic membrane pore-forming toxins. Structure. 2011, 19(2): 181-191.
- Tanaka K, et al. Structural basis for self-assembly of a cytolytic pore lined by protein and lipid. Nature Communications. 2015, 6(1): 6337.
- Song C, et al. Crystal structure and functional mechanism of a human antimicrobial membrane channel. Proceedings of the National Academy of Sciences. 2013, 110(12): 4586-4591.
- Meusch D, et al. Mechanism of Tc toxin action revealed in molecular detail. Nature. 2014, 508(7494): 61-65.
- Gatsogiannis C, et al. Membrane insertion of a Tc toxin in near-atomic detail. Nature Structural & Molecular Biology. 2016, 23(10): 884-890.
- Roderer D, et al. Structure of a Tc holotoxin pore provides insights into the translocation mechanism. Proceedings of the National Academy of Sciences. 2019, 116(46): 23083-23090.
- Köster S, et al. Crystal structure of listeriolysin O reveals molecular details of oligomerization and pore formation. Nature Communications. 2014, 5(1): 3690.
- Schubert E, et al. Membrane insertion of α-xenorhabdolysin in near-atomic detail. Elife. 2018, 7: e38017.
- Zhang K, et al. Cryo-EM structures of Helicobacter pylori vacuolating cytotoxin A oligomeric assemblies at near-atomic resolution. Proceedings of the National Academy of Sciences. 2019, 116(14): 6800-6805.
- Su M, et al. Cryo-EM analysis reveals structural basis of Helicobacter pylori VacA toxin oligomerization. Journal of Molecular Biology. 2019, 431(10): 1956-1965.
- Schureck M A, et al. Malaria parasites use a soluble RhopH complex for erythrocyte invasion and an integral form for nutrient uptake. Elife. 2021, 10: e65282.
- Byrne M J, et al. Cryo-EM structures of an insecticidal Bt toxin reveal its mechanism of action on the membrane. Nature Communications. 2021, 12(1): 2791.