Structural Research of Lipoxygenases
Lipoxygenases (EC1.13.11. -) is a structurally related non-heme iron dioxygenases family. Lipoxygenases (LOX) play a role in the production and, in some cases, the metabolism of fatty acid hydroperoxides. The LOX family is involved in the biosynthesis of various lipid mediators, including leukotrienes and lipotoxins, which regulate inflammatory and immune responses.
X-ray crystallography is an essential tool for analyzing complex biomolecules, which can reveal the three-dimensional structure and function of LOX. This technology has successfully described the crystal structure of LOX in plants, animals, bacteria, and fungi. These data provide essential information for studying the mechanism of LOX catalytic reactions and protein-protein small molecule interactions. In addition, X-ray crystallography can also be used to explore the structure and function of many other complex biomolecules, such as membrane proteins, enzymes, antibodies, etc.
For example, scientists obtained stable 5-LOX by replacing a sequence of the carboxyl end involved in directed binding, catalytic iron in 5-LOX. Then, the scientists reported the crystal structure of stable human 5-lipoxygenase at a resolution of 2.4 Å.
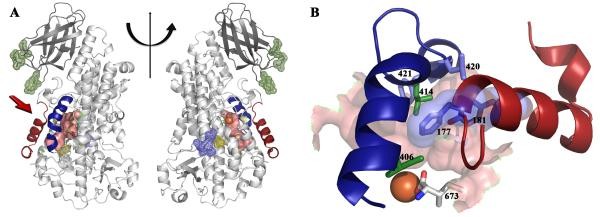 Figure 1.
The structure of Stable-5-LOX (GILBERT et al., 2011)
Figure 1.
The structure of Stable-5-LOX (GILBERT et al., 2011)
The function of LOX in microorganisms is not yet clear, but it is suspected to be related to host-pathogen interactions. In 2019, researchers reported the crystal structures of five types of LOX from Fusarium graminearum (Fg). A single FgLOX monomer consists of 29 α-helices. All but two of these parts gather together to form a flat triangular bundle. This LOX exhibits an amino-terminal extension different from that of animals and plants, providing an enveloping domain for dimerization and seemingly interfering with the Fe coordination sphere, as no typical LOX configuration was observed at the catalytic metal site when the enzyme is a dimer.
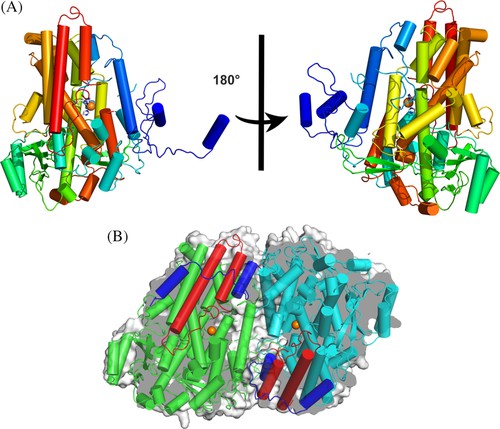 Figure 2. Dimeric FgLOX
(PAKHOMOVA, et al., 2019)
Figure 2. Dimeric FgLOX
(PAKHOMOVA, et al., 2019)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Arachidonic acid 15-lipoxygenase from soybeans | Glycine max | X-ray diffraction | 2.60 Å | 2SBL |
| Lipoxygenase-L1 from soybeans | Glycine max | X-ray diffraction | 1.40 Å | 1YGE |
| Lipoxygenase-L3 from soybeans | Glycine max | X-ray diffraction | 2.60 Å | 1LNH |
| 8R-Lipoxygenase from coral | Plexaura homomalla | X-ray diffraction | 3.20 Å | 2FNQ |
| 8R-Lipoxygenase from coral. Δ413-417 I805A | Plexaura homomalla | X-ray diffraction | 1.85 Å | 3FG4 |
| 15-Lipoxygenase from rabbit reticulocytes | Oryctolagus cuniculus | X-ray diffraction | 2.40 Å | 1LOX |
| 5-Lipoxygenase | Homo sapiens | X-ray diffraction | 2.39 Å | 3O8Y |
| 15-Lipoxygenase-2 (15-LOX-2) with substrate mimic | Homo sapiens | X-ray diffraction | 2.63 Å | 4NRE |
Table 1. Structural research of lipoxygenases.
Creative Biostructure focuses on providing high-quality X-ray crystallography services for life sciences. X-ray crystallography technology can reveal the three-dimensional structure and function of biological macromolecules and is an important tool for analyzing complex biological molecules such as membrane proteins.
We use high-strength X-rays and precise detectors to obtain high-resolution structural data, and we are committed to providing customers with the best service. Whether you are in drug development, genetic engineering, biotechnology, or other life sciences, we can provide you with the best quality X-ray crystallography services to help you understand the structure and function of biomolecules in depth and support your research.
References
- KOBE M J, et al. The structure of human 15-lipoxygenase-2 with a substrate mimic. Journal of Biological Chemistry, 2014, 289(12): 8562–8569.
- GILBERT N C, et al. The structure of human 5-lipoxygenase. Science, 2011, 331(6014): 217–219.
- GILLMOR S A, et al. The structure of mammalian 15-lipoxygenase reveals similarity to the lipases and the determinants of substrate specificity. Nature Structural Biology, 1997, 4(12): 1003–1009.
- NEAU D B, et al. The 1.85 Å structure of an 8r-lipoxygenase suggests a general model for lipoxygenase product specificity. Biochemistry, 2009, 48(33): 7906–7915.
- OLDHAM M L, et al. Insights from the X-ray crystal structure of Coral 8R-lipoxygenase. Journal of Biological Chemistry, 2005, 280(47): 39545–39552.
- SKRZYPCZAK-JANKUN E, et al. Structure of soybean lipoxygenase L3 and a comparison with its L1 isoenzyme. Proteins: Structure, Function, and Genetics, 1997, 29(1): 15–31.
- MINOR W, et al. Crystal structure of soybean lipoxygenase L-1 at 1.4 Å resolution,. Biochemistry, 1996, 35(33): 10687–10701.
- BOYINGTON J C, et al. The three-dimensional structure of an arachidonic acid 15-lipoxygenase. Science, 1993, 260(5113): 1482–1486.
- PAKHOMOVA S, et al. An ensemble of lipoxygenase structures reveals novel conformations of the Fe Coordination Sphere. Protein Science, 2019, 28(5): 920–927.