Structural Research of Transhydrogenases
Transhydrogenases (TH) are widely present in organisms and play essential roles in maintaining cellular redox homeostasis with implications for aging and many human diseases. Two types of TH have been found to exist in aerobic organisms, membrane-bound proton-pumping NADH: NADP+ oxidoreductase (PntAB) and membrane-independent soluble transhydrogenase (UdhA).
Advances in research on the structure of TH
TH exists as homodimers, each containing a proton translocation transmembrane structural domain and two soluble nucleotide-binding structural domains that mediate hydride transfer between NAD(H) and NADP(H). The three-domain structure of TH is conserved across species, but the polypeptide composition varies considerably. Researchers have analyzed the crystal structure of TH by single-particle cryo-electron microscopy, which helps to understand its role under pathological conditions, making it a potential target for new therapeutic strategies for oxidative stress-mediated diseases and cancer.
Molecular structure analysis of StnABC
StnABC acts as an electron-divergent transhydrogenase to catalyze reduction-dependent reactions. The researchers revealed the molecular structure of StnABC from S. ovata and observed that the complex consists of four protomers, each a StnABC heterotrimer. The tetrameric core consists of the StnC subunit of each protomer flanked by the more energetic StnAB subcomplexes, with only StnB binding to StnC, whereas StnA is found to interact with StnB through multiple contacts. The entire complex contains a large number of redox cofactors, and it is evident from the structure that the distances from all the iron-sulfur clusters to the sides of a single proto membrane ensure rapid electron transfer throughout the structure.
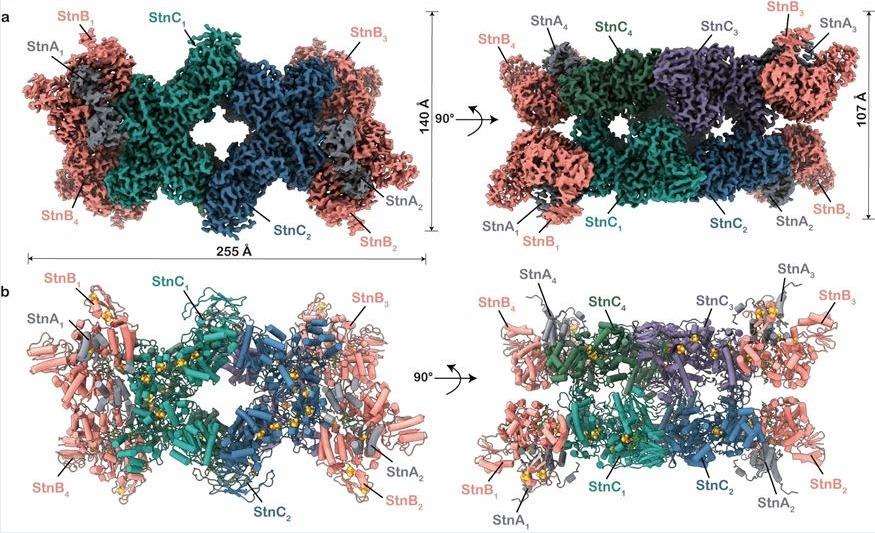 Figure 1. Molecular structure of StnABC. (Kumar A, et al., 2023)
Figure 1. Molecular structure of StnABC. (Kumar A, et al., 2023)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Transhydrogeanse domain II dimer | Thermus thermophilus HB27 | X-ray diffraction | 2.77 Å | 4O93 |
| Membrane-bound transhydrogenase | Thermus thermophilus | X-ray diffraction | 2.2 Å | 5UNI |
| Transhydrogenase in the apo state | Ovis aries | Cryo-EM single particle analysis | 3.7 Å | 6S59 |
| Transhydrogenase domain I | Escherichia coli | X-ray diffraction | 1.9 Å | 1X13 |
| Transhydrogenase domain I with bound NAD | Escherichia coli | X-ray diffraction | 1.94 Å | 1X14 |
| Transhydrogenase domain I with bound NADH | Escherichia coli | X-ray diffraction | 2.04 Å | 1X15 |
| Transhydrogenase dIII bound with NADPH | Homo sapiens | X-ray diffraction | 2.2 Å | 1U31 |
| Transhydrogenase in the presence of NADP+ in a "single face-down" conformation | Ovis aries | Cryo-EM single particle analysis | 3.7 Å | 6QUE |
| Transhydrogenase in the presence of NADP+ in a "double face-down" conformation | Ovis aries | Cryo-EM single particle analysis | 2.9 Å | 6QTI |
| Transhydrogenase domain III | Bos taurus | X-ray diffraction | 1.21 Å | 1D4O |
| Transhydrogenase coupling proton translocation and hydride transfer | Thermus thermophilus HB27 | X-ray diffraction | 6.926 Å | 4O9U |
| Transhydrogenase coupling proton translocation and hydride transfer | Thermus thermophilus HB27 | X-ray diffraction | 3.079 Å | 4O9T |
| Transhydrogeanse domain II dimer SeMet derivative | Thermus thermophilus HB27 | X-ray diffraction | 1.89 Å | 4OP9 |
| The electron bifurcating transhydrogenase StnABC complex (state 1) | Sporomusa ovata DSM 2662 | Cryo-EM single particle analysis | 3.2 Å | 8OH9 |
| The electron bifurcating transhydrogenase StnABC complex (state 2) | Sporomusa ovata DSM 2662 | Cryo-EM single particle analysis | 3 Å | 8OH5 |
| Transhydrogenase domain III bound to NADPH | Rhodospirillum rubrum | X-ray diffraction | 2.4 Å | 1PNQ |
| Transhydrogenase domain I without bound NAD(H) | Rhodospirillum rubrum | X-ray diffraction | 1.81 Å | 1L7D |
| Transhydrogenase domain I with bound NADH | Rhodospirillum rubrum | X-ray diffraction | 1.9 Å | 1L7E |
| Transhydrogenase domain III bound to NADP | Rhodospirillum rubrum | X-ray diffraction | 2.1 Å | 1PNO |
| Transhydrogenase [(domain I)2: domain III] heterotrimer complex | Rhodospirillum rubrum | X-ray diffraction | 3.1 Å | 1XLT |
| Transhydrogenase (dI. Q132N)2(dIII)1 asymmetric complex | Rhodospirillum rubrum | X-ray diffraction | 2.4 Å | 1NM5 |
| Transhydrogenase (dI. NADH)2(dIII.NADPH)1 asymmetric complex | Rhodospirillum rubrum | X-ray diffraction | 3 Å | 1U2D |
| Transhydrogenase (dI. R127A.NAD+)2(dIII.NADP+)1 asymmetric complex | Rhodospirillum rubrum | X-ray diffraction | 2.6 Å | 2FR8 |
| Transhydrogenase (dI. S138A. NADH)2(dIII.NADPH)1 asymmetric complex | Rhodospirillum rubrum | X-ray diffraction | 3.2 Å | 2FRD |
| Transhydrogenase (dI.ADPr)2(dIII.NADPH)1 asymmetric complex | Rhodospirillum rubrum | X-ray diffraction | 2.2 Å | 1U2G |
| Transhydrogenase (dI.H2NADH)2(dIII.NADP+)1 asymmetric complex | Rhodospirillum rubrum | X-ray diffraction | 2.6 Å | 2OO5 |
| DI and DIII complex of transhydrogenase with a thio-nicotinamide nucleotide analogue | Rhodospirillum rubrum | X-ray diffraction | 2.61 Å | 1PTJ |
Table 1. Structural research of the transhydrogenases.
Creative Biostructure is committed to providing high-quality structural analysis services and expertise to help our clients advance their research and achieve their scientific goals. Our experienced scientists are skilled in X-ray crystallography, cryo-electron microscopy (cryo-EM), nuclear magnetic resonance (NMR), and other structural biology techniques. We also provide additional services such as functional determination and mechanistic analysis of transhydrogenases to provide a comprehensive understanding of the structure and function of the target protein.
If you are interested in learning more about our structural analysis services for transhydrogenases or other membrane proteins, please feel free to contact us. Our experts will be pleased to discuss your project and provide a customized solution that meets your requirements.
References
- Kumar A, et al. Molecular architecture and electron transfer pathway of the Stn family transhydrogenase. Nat Commun. 2023. 14(1): 5484.
- Zhang Q, et al. Proton-translocating nicotinamide nucleotide transhydrogenase: a structural perspective. Front Physiol. 2017. 8: 1089.
- Francisco A, et al. Mitochondrial NAD(P)+ Transhydrogenase: From Molecular Features to Physiology and Disease. Antioxid Redox Signal. 2022. 36(13-15): 864-884.