Structural Research of O-acyltransferases
Membrane-bound O-acyltransferase (MBOAT) is involved in neutral lipid biosynthesis, protein/peptide acylation, and phospholipid remodeling. The members of this family all contain multiple transmembrane domains, and most carry two conserved residues, one conserved histidine (His) embedded in a hydrophobic residue segment and one asparagine (Asn) or histidine located in a more hydrophilic upstream region. Some MBOAT members are drug targets for human diseases, including atherosclerosis, obesity, Alzheimer's disease, and viral infection.
Structural research on the bacterial DltB protein
The structure of DltB, an MBOAT protein from Streptococcus thermophilus (Q5M4V4), has been investigated at 3.3 Å resolution. DltB is essential for D-alanylation of bacterial cell wall phosphomimetic acids. It contains 415 amino acid residues, with both the N- and C-termini located extracellularly. It forms 17 helices located mainly within the lipid bilayer. Of these, 11 transmembrane helices form an external circular ridge and shield the thinner central region. This region is the core site involved in catalysis.
Action mechanism of DltB protein
There is an approximately straight tunnel between the bottom of the extracellular funnel and the cytoplasmic side. The tunnel is formed by three DltB spirals (H13-H15) and a horizontal spiral H12 at the center core. The residues in the tunnel are highly conserved in the DltB protein. This tunnel is connected to partner DltC, which carries D-alanine to the bottom of the funnel for catalytic reactions.
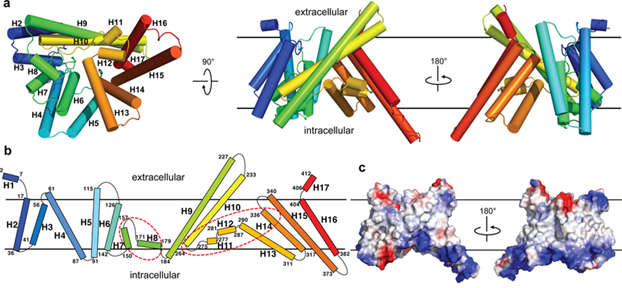 Figure 1. The overall structure of DltB. (Ma D, et al., 2018)
Figure 1. The overall structure of DltB. (Ma D, et al., 2018)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| A membrane protein, crystal form I | Streptococcus thermophilus | X-ray diffraction | 3.27 Å | 6BUG |
| A membrane protein, crystal form II | Streptococcus thermophilus | X-ray diffraction | 3.15 Å | 6BUH |
| A membrane protein, crystal form III | Streptococcus thermophilus | X-ray diffraction | 3.27 Å | 6BUI |
| Diacylglycerol Acyltransferase 1 in complex with oleoyl-CoA | Homo sapiens | Cryo-EM single particle analysis | 3.1 Å | 6VP0 |
| Sterol O-acyltransferase 1 in complex with CI-976 | Homo sapiens | Cryo-EM single particle analysis | 3.5 Å | 6L47 |
| Sterol O-acyltransferase 1 in resting state | Homo sapiens | Cryo-EM single particle analysis | 3.5 Å | 6L48 |
| Dimeric of ACAT1 | Homo sapiens | Cryo-EM single particle analysis | 3 Å | 6P2J |
| Tetrameric of ACAT1 | Homo sapiens | Cryo-EM single particle analysis | 3.1 Å | 6P2P |
| Nevanimibe bound ACAT2 | Homo sapiens | Cryo-EM single particle analysis | 3.93 Å | 7N6R |
| Triacylglycerol synthesizing enzyme DGAT1 in complex with T863 inhibitor | Homo sapiens | Cryo-EM single particle analysis | 3.2 Å | 8ESM |
| Triacylglycerol synthesizing enzyme DGAT1 in complex with DGAT1IN1 inhibitor | Homo sapiens | Cryo-EM single particle analysis | 3.2 Å | 8ETM |
| Diacylglycerol O-acyltransferase 1 | Homo sapiens | Cryo-EM single particle analysis | 3 Å | 6VYI |
| Diacylglycerol O-acyltransferase 1 complexed with acyl-CoA substrate | Homo sapiens | Cryo-EM single particle analysis | 3.2 Å | 6VZI |
| Nevanimibe-bound tetrameric ACAT1 | Homo sapiens | Cryo-EM single particle analysis | 3.67 Å | 6VUM |
| PPPA boundACAT2 | Homo sapiens | Cryo-EM single particle analysis | 3.87 Å | 7N6Q |
| Hedgehog acyltransferase (HHAT) in complex with palmitoyl-CoA and two Fab antibody fragments | Homo sapiens | Cryo-EM single particle analysis | 2.7 Å | 7MHY |
| Hedgehog acyltransferase (HHAT) in complex with megabody 177 bound to non-hydrolysable palmitoyl-CoA (Composite Map) | Homo sapiens | Cryo-EM single particle analysis | 2.7 Å | 7Q1U |
| Hedgehog acyltransferase (HHAT) in complex with megabody 177 bound to IMP-1575 | Homo sapiens | Cryo-EM single particle analysis | 3.59 Å | 7Q6Z |
| PORCN in complex with Palmitoleoyl-CoA | Homo sapiens | Cryo-EM single particle analysis | 3.11 Å | 7URA |
| PORCN in complex with LGK974 | Homo sapiens | Cryo-EM single particle analysis | 3.14 Å | 7URC |
| PORCN in complex with palmitoleoylated WNT3A peptide | Homo sapiens | Cryo-EM single particle analysis | 3.19 Å | 7URE |
| PORCN in complex with LGK974 and WNT3A peptide | Homo sapiens | Cryo-EM single particle analysis | 2.92 Å | 7URD |
| Lysophospholipid acyltransferase LPCAT3 (MOBAT5) in its monomeric and apo form | Gallus gallus | X-ray diffraction | 3.4 Å | 7EWT |
| Lysophospholipid acyltransferase LPCAT3 in complex with lysophosphatidylcholine | Gallus gallus | Cryo-EM single particle analysis | 3.57 Å | 7F3X |
| Brassica napus DGAT1 exosite | Brassica napus | SOLUTION NMR | / | 5UZL |
Table 1. Structural research of membrane-bound O-acyltransferase (MBOAT).
Creative Biostructure has long been committed to the study of structural biology and membrane proteins. We have extensive experience in determining membrane protein structures using NMR spectroscopy, cryo-electron microscopy (cryo-EM) and X-ray crystallography. Studying the structure of MBOAT family proteins is beneficial for the development of antibacterial agents and the investigation of drug targets for human diseases.
In addition to the structural determination of membrane proteins, we can accurately analyze biomolecules, including but not limited to nucleic acids, ribosomes, small proteins, protein complexes, protein-ligand complexes, and viruses. If you are interested in our services, please contact us for more details.
References
- Ma D, et al. Crystal structure of a membrane-bound O-acyltransferase. Nature. 2018. 562(7726):286-290.
- Chang, et al. Membrane-bound O-acyltransferases (MBOATs). Frontiers in Biology 6. 2011:177-182.