Structural Research of Outer Membrane Proteins
Outer Membrane Protein (OMP) has an all-β chain transmembrane structure, and its number and diversity are lower than that of all-α helical transmembrane proteins. However, as outer membrane channels, OMPs are responsible for the transport of substrates, ions, and even small molecules, and are of great significance in antibiotic resistance, vaccine design, and cancer diagnosis and treatment. Therefore, the study of outer membrane proteins is an indispensable step for further insight into the microscopic activities and the mechanism of life.
OMP anatomy
The β-barrel chains of OMP are amphiphilic and usually inversely parallel to each other. The structure of the chains bound through the hydrogen of the main chain results in the side chains alternating between pore-facing and membrane-facing side chains. Rings connect the chains, with larger rings located in the extracellular space and smaller ones in the periplasmic space. The β-barrels of most OMPs are monomeric, with one chain forming a barrel. However, some barrels undergo oligomerization, most commonly forming trimers.
Structural studies on the β-barrel assembly machinery (BAM) of Gram-negative bacteria
Assembly of β-OMP in Gram-negative bacteria is mediated by β-barrel assembly machinery (BAM) complexes. Using a detergent-free system containing styrene-maleic acid (SMA) copolymer, the BAM-MBP-76EspP co-composite is directly dissolved and separated into natural nanodisks. The structure is resolved to a global resolution of 3.6 Å by single-particle cryoelectron microscopy. BAM is found to bind the highly conserved "β-signaling" motif of EspP, which is correctly localized in the OM during folding. Four C-terminal β-strands actively folding the β-barrel of EspP can be distinguished, and these β-strands extend into the low-resolution region of the OM nanodisks.
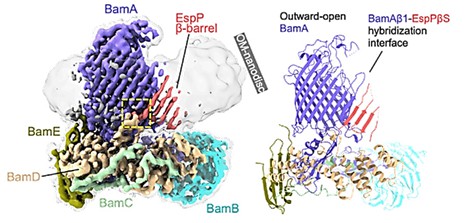 Figure 1. High-resolution cryo-EM map (left, 3.6 Å) and BAM-MBP-76EspP model (Right). (Doyle MT, et al., 2022)
Figure 1. High-resolution cryo-EM map (left, 3.6 Å) and BAM-MBP-76EspP model (Right). (Doyle MT, et al., 2022)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| BAM complex | Escherichia coli K-12 | X-ray diffraction | 3.28 Å | 6LYR |
| BAM complex | Escherichia coli K-12 | X-ray diffraction | 3.05 Å | 6LYS |
| BAM complex | Escherichia coli K-12 | Cryo-EM single particle analysis | 4.2 Å | 6LYU |
| Outer membrane protein a (OmpA) transmembrane domain | Escherichia coli BL21(DE3) | X-ray diffraction | 2.5 Å | 1BXW |
| Outer membrane protein OprG | Pseudomonas aeruginosa | X-ray diffraction | 2.4 Å | 2X27 |
| A membrane complex | Escherichia coli K-12 | X-ray diffraction | 3.555 Å | 5AYW |
| A membrane protein | Pseudomonas aeruginosa PAO1 | X-ray diffraction | 3.1 Å | 5AZS |
| BamACDE complex | Escherichia coli | X-ray diffraction | 3.5 Å | 5D0Q |
| BamABCDE complex | Escherichia coli | X-ray diffraction | 2.9 Å | 5D0O |
| The drug-discharge outer membrane protein, OprM | Pseudomonas aeruginosa | X-ray diffraction | 2.56 Å | 1WP1 |
| The outer membrane protein a (OmpA) transmembrane domain | Escherichia coli BL21(DE3) | X-ray diffraction | 1.65Å | 1QJP |
| The outer membrane protein OmpX | Escherichia coli | X-ray diffraction | 1.9 Å | 1QJ8 |
| BAM-EspP complex structure with BamA-G431C and G781C/EspP-N1293C and A1043C mutations in nanodisc | Escherichia coli K-12 Escherichia coli O157:H7 |
Cryo-EM single particle analysis | 3.4 Å | 7YE4 |
| BAM-EspP complex structure with BamA-N427C/EspP-R1297C mutations in nanodisc | Escherichia coli K-12 Escherichia coli O157:H7 |
Cryo-EM single particle analysis | 3.4 Å | 7YE6 |
| BAM-EspP complex structure with BamA-S425C/EspP-S1299C mutations in nanodisc | Escherichia coli K-12 | Cryo-EM single particle analysis | 3.1 Å | 8BO2 |
| BamACDE subcomplex | Escherichia coli K-12 | X-ray diffraction | 3.392 Å | 5EKQ |
| The darobactin-bound BAM complex (BamABCDE) | Escherichia coli K-12 synthetic construct |
X-ray diffraction | 3.03 Å | 7NRI |
| Outer membrane protein a transmembrane domain: 10 conformers | Escherichia coli | SOLUTION NMR | / | 1G90 |
| Outer membrane protein A transmembrane domain | Escherichia coli | SOLUTION NMR | / | 2GE4 |
| BAM in MSP1E3D1 nanodiscs | Escherichia coli | Cryo-EM single particle analysis | 5.9 Å | 7RI6 |
| A substrate-engaged Bam complex | Escherichia coli K-12 | X-ray diffraction | 4.1 Å | 6V05 |
| A BAM in MSP1E3D1 nanodiscs at 4 Angstrom resolution | Escherichia coli | Cryo-EM single particle analysis | 4 Å | 7RI5 |
| Lateral-closed conformation of the lid-locked BAM complex | Escherichia coli K-12 | Cryo-EM single particle analysis | 4.1 Å | 7BNQ |
| A minimal transmembrane beta-barrel platform protein | Escherichia coli | SOLUTION NMR | / | 2JMM |
| The N-terminal beta-barrel domain of OprF | Pseudomonas aeruginosa | X-ray diffraction | 1.6 Å | 4RLC |
Table 1. Structural research of outer membrane proteins.
Creative Biostructure has long been committed to the study of structural biology and membrane proteins. We provide NMR spectroscopy, cryo-electron microscopy (cryo-EM) and X-ray crystallography to assist clients in studying outer membrane protein structures and to further understand the transport mechanism of outer membranes, which with important roles in antibiotic resistance, vaccine design, and cancer diagnosis and treatment.
Our experts have extensive experience in determining membrane protein structures. If you are interested in our services, please contact us for more details.
References
- Doyle MT, et al. Cryo-EM structures reveal multiple stages of bacterial outer membrane protein folding.Cell. 2022.185(7):1143-1156.
- Slusky JS. Outer membrane protein design. Curr Opin Struct Biol. 2017. (45):45-52.