Structural Research of Beta-Barrel Membrane Proteins: Porins
Beta-barrel membrane proteins are a group of proteins that form pores in the outer membrane of Gram-negative bacteria, mitochondria, and chloroplasts. Porins are a subclass of beta-barrel membrane proteins that function as channels for the transport of small molecules such as nutrients, ions, and antibiotics across the bacterial outer membrane.
Porins are typically composed of 16-18 antiparallel beta-strands that form a barrel-like structure. The strands are connected by short turns or loops that are located on the periplasmic side of the barrel. Porins are asymmetric and have a hydrophilic lumen that allows for the diffusion of small molecules, while the exterior surface is hydrophobic and interacts with the lipid bilayer. The porin structure is essential for its function as a molecular sieve and for its interaction with other molecules.
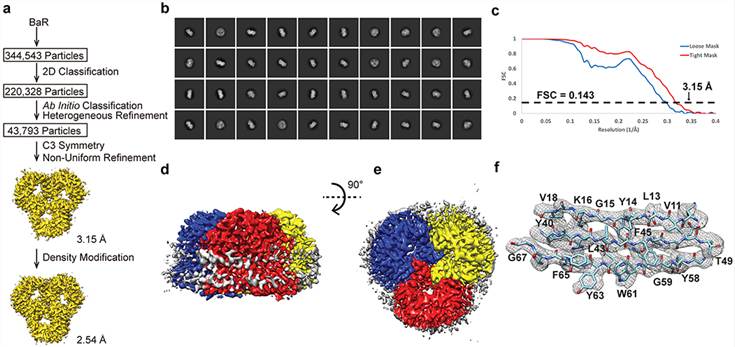 Figure 1. Cryo-EM analysis of the E. coli OmpF porin channel. (Su C C, et al., 2021)
Figure 1. Cryo-EM analysis of the E. coli OmpF porin channel. (Su C C, et al., 2021)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Porin | Rhodobacter capsulatus | X-ray diffraction | 1.80 Å | 2POR |
| Porin | Fuscovulum blasticum | X-ray diffraction | 1.96 Å | 1PRN |
| Porin, E1M/A116K Mutant (expressed in E. coli) | Fuscovulum blasticum | X-ray diffraction | 2.19 Å | 1BH3 |
| Porin (expressed in E. coli) | Fuscovulum blasticum | X-ray diffraction | 3.00 Å | 1H6S |
| OmpK36 osmoporin (expressed in E. coli) | Klebsiella pneumoniae | X-ray diffraction | 3.20 Å | 1OSM |
| Wild type OmpK36 (expressed in E. coli) | Klebsiella pneumoniae | X-ray diffraction | 1.98 Å | 6RD3 |
| Omp32 anion-selective porin | Comamonas acidovorans | X-ray diffraction | 2.10 Å | 1E54 |
| Omp32 anion-selective porin | Delftia acidovorans | X-ray diffraction | 1.50 Å | 2FGR |
| OmpF Porin | Escherichia coli | X-ray diffraction | 2.40 Å | 2OMF |
| OmpF Porin from colicin-resistant E. coli | Escherichia coli | X-ray diffraction | 3.00 Å | 1MPF |
| OmpF Porin | Escherichia coli | X-ray diffraction | 3.20 Å | 1OPF |
| OmpF Porin, D113G mutant (expressed in E. coli) | Escherichia coli | X-ray diffraction | 3.50 Å | 1GFM |
| OmpF Porin, D74A mutant (expressed in E. coli) | Escherichia coli | X-ray diffraction | 3.00 Å | 1BT9 |
| OmpF Porin, Y106F Mutant (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.20 Å | 1HXX |
| OmpF Porin (expressed in E. coli) | Escherichia coli | X-ray diffraction | 1.59 Å | 2ZFG |
| OmpF porin with a synthetic dibenzo-18-crown-6 (expressed in E. coli) | Escherichia coli | X-ray diffraction | 3.40 Å | 3FYX |
| OmpF in complex with colicin peptide OBS1 | Escherichia coli | X-ray diffraction | 3.01 Å | 3O0E |
| OmpF porin (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.61 Å | 3HW9 |
| OmpF Porin in presence of foscholine-12 | Escherichia coli | X-ray diffraction | 3.79 Å | 3K19 |
| OmpF porin in lipidic cubic phase | Escherichia coli | X-ray diffraction | 2.80 Å | 3POU |
| OmpF Porin | Escherichia coli | X-ray diffraction | 3.50 Å | 4D5U |
| OmpF Porin | Escherichia coli | Cryo-EM single particle analysis | 3.15 Å | 6WTZ |
| OmpF and BtuB Porin in complex with colicin E9 (colE9) (expressed in E. coli) | Escherichia coli | Cryo-EM single particle analysis | 4.70 Å | 7NSU |
| Omp35 (OmpF) (expressed in E. coli) | Klebsiella aerogenes | X-ray diffraction | 2.85 Å | 5O78 |
| OmpF Porin (expressed in E. coli) | Salmonella enterica | X-ray diffraction | 2.79 Å | 3NSG |
| OmpE35 (OmpF) (expressed in E. coli) | Enterobacter cloacae | X-ray diffraction | 2.30 Å | 6ENE |
| OmpE35 (OmpF) (expressed in E. coli) | Klebsiella pneumoniae | X-ray diffraction | 1.50 Å | 5O77 |
| OmpC Osmoporin (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.00 Å | 2J1N |
| OmpC Osmoporin clinical variant OmpC06 (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.50 Å | 2XE1 |
| OmpC Osmoporin | Escherichia coli | Cryo-EM single particle analysis | 2.56 Å | 7JZ3 |
| OmpK36 (OmpC) (expressed in E. coli) | Klebsiella pneumoniae | X-ray diffraction | 1.65 Å | 5O79 |
| Omp36 (OmpC) (expressed in E. coli) | Klebsiella aerogenes | X-ray diffraction | 2.47 Å | 5O9C |
| OmpC homolog (OmpE36) with bound lipopolysaccharide (LPS) (expressed in E. coli) | Enterobacter cloacae | X-ray diffraction | 1.45 Å | 5FVN |
| Monomeric porin OmpG (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.30 Å | 2F1C |
| OmpG *monomeric* porin in open state (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.30 Å | 2IWV |
| OmpG *monomeric* porin (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.18 Å | 2X9K |
| OmpG *monomeric* porin (expressed in E. coli) | Escherichia coli | Solution NMR | / | 2JQY |
| OmpG *monomeric* porin (expressed in E. coli) | Escherichia coli | Solid-state NMR | / | 5MWV |
| OmpG ΔL6-ΔD215 mutant, "quiet" porin (expressed in E. coli) | Escherichia coli | Solution NMR | / | 6OQH |
| OmpT porin (outer membrane expressed) (expressed in E. coli) | Vibrio cholerae | X-ray diffraction | 3.20 Å | 5OYK |
| Trimeric OmpU structure (expressed in E. coli) | Vibrio cholerae | X-ray diffraction | 2.22 Å | 5ONU |
| OmpU porin (expressed in E. coli) | Vibrio cholerae | X-ray diffraction | 1.55 Å | 6EHB |
| PhoE | Escherichia coli | X-ray diffraction | 3.00 Å | 1PHO |
| LamB Maltoporin | Salmonella enterica | X-ray diffraction | 2.40 Å | 2MPR |
| LamB Maltoporin | Escherichia coli | X-ray diffraction | 3.10 Å | 1MAL |
| LamB Maltoporin in complex with maltose | Escherichia coli | X-ray diffraction | 2.60 Å | 1MPM |
| LamB Maltoporin in complex with sucrose (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.40 Å | 1AF6 |
| ScrY sucrose-specific porin (expressed in E. coli) | Salmonella enterica | X-ray diffraction | 2.40 Å | 1A0T |
| MspA mycobacterial porin (expressed in E. coli) | Mycolicibacterium smegmatis | X-ray diffraction | 2.50 Å | 1UUN |
| OprB carbohydrate-specific transporter at high pH (expressed in E. coli) | Pseudomonas putida | X-ray diffraction | 2.70 Å | 4GEY |
| OprO diphosphate-specific transporter (expressed in E. coli) | Pseudomonas aeruginosa | X-ray diffraction | 1.52 Å | 4RJW |
| OprP phosphate-specific transporter | Pseudomonas aeruginosa | X-ray diffraction | 1.94 Å | 2O4V |
| PorB outer membrane protein, native structure (expressed in E. coli) | Neisseria meningitidis | X-ray diffraction | 2.30 Å | 3VZT |
| Wild-type PorB (expressed in E. coli) | Neisseria meningitidis | X-ray diffraction | 3.32 Å | 3WI4 |
| PorB outer membrane protein, G103K mutant (expressed in E. coli) | Neisseria meningitidis | X-ray diffraction | 2.76 Å | 7DE8 |
| PorB outer membrane protein | Neisseria gonorrhoeae | X-ray diffraction | 3.20 Å | 4AUI |
| KdgM *monomeric* porin in complex with disordered oligogalacturonate, wild-type (expressed in E. coli) | Dickeya dadantii | X-ray diffraction | 1.93 Å | 4FQE |
| CymA monomeric outer membrane protein (NHis-SeMet) (expressed in E. coli) | Klebsiella oxytoca | X-ray diffraction | 2.51 Å | 4V3G |
| COG4313 outer membrane channel (expressed in E. coli) | Pseudomonas putida | X-ray diffraction | 2.30 Å | 4RL8 |
| OprG outer membrane amino acid transporter (expressed in E. coli) | Pseudomonas aeruginosa | Solution NMR | / | 2N6L |
| MOMP major outer membrane protein (expressed in E. coli) | Campylobacter jejuni | X-ray diffraction | 2.10 Å | 5LDV |
| Omp-Pst1 type-A porin (expressed in E. coli) | Providencia stuartii | X-ray diffraction | 3.20 Å | 4D64 |
| Chitoporin (ChiP), in vitro-folded crystal form I (expressed in E. coli) | Vibrio harveyi | X-ray diffraction | 1.95 Å | 5MDO |
| FapF amyloid secretion protein FapFβ (expressed in E. coli) | Pseudomonas sp. UK4 | X-ray diffraction | 2.50 Å | 5O65 |
| DcaP outer membrane channel (expressed in E. coli) | Acinetobacter baumannii | X-ray diffraction | 2.20 Å | 6EUS |
| MtrAB electron transporter complex | Shewanella baltica | X-ray diffraction | 2.70 Å | 6R2Q |
Table 1. Structural Research of Beta-Barrel Membrane Proteins: Porins.
Structural biology services offered by Creative Biostructure can help researchers in the exploration of porin structures. We provide custom protein expression, purification, and crystallization services, as well as X-ray crystallography and cryo-EM services for structure determination. Our team has years of experience in the field of structural biology and can provide high-quality services tailored to specific research needs.
The exploration of porin structures is a field that requires a vast amount of knowledge and expertise. If you are intrigued by the complexities of these intricate membrane proteins and would like to delve deeper into the science behind them, our team at Creative Biostructure is here to assist you. We offer an array of structural biology services that cater to the diverse needs of the scientific community, and we take great pride in providing our clients with the highest level of quality and efficiency. We encourage you to contact us today to learn more about our comprehensive range of services and how we can help you achieve your research goals.
References
- Su C C, et al. A 'Build and Retrieve' methodology to simultaneously solve cryo-EM structures of membrane proteins. Nature Methods. 2021, 18(1): 69-75.
- Edwards M J, et al. The crystal structure of a biological insulated transmembrane molecular wire. Cell. 2020, 181(3): 665-673. e10.