Structural Research of Bacterial Mercury Detoxification Proteins
Mercury (Hg) is a toxic heavy metal that poses a significant threat to human health and the environment. Bacteria have evolved a variety of mechanisms to detoxify Hg, including the expression of specific proteins that bind and sequester Hg ions. Structural research on these bacterial Hg detoxification proteins is crucial for understanding their mechanisms of action and developing new strategies for the remediation of Hg-contaminated sites.
In the bacterial mercury detoxification system, Hg (II) is transported across the intracellular membrane into the cytoplasm by membrane proteins such as MerF. With both metal transfer and transport activities, the structural biology of MerF is too complex to be explained simply by the binding of Hg (II) to cysteine pairs. Therefore, it is necessary to determine the three-dimensional structure in which it performs its function in the phospholipid bilayer. MerFt is the 60-residue helix-loop-helix integral membrane core of MerF. Some researchers have studied the three-dimensional structure of MerFt in proteoliposomes by using the NMR method.
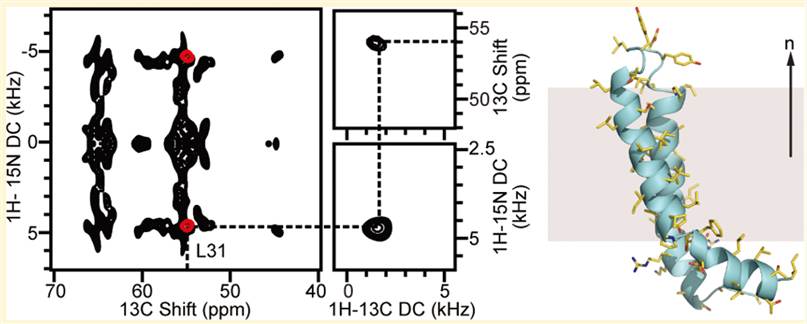 Figure 1. Structure of MerFt in DMPC phospholipid bilayers. (Das B B, et al., 2012)
Figure 1. Structure of MerFt in DMPC phospholipid bilayers. (Das B B, et al., 2012)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| MerF Hg (II) transporter, truncated protein (AAs 13-72) determined in aligned bicelles (expressed in E. coli) | Morganella morganii | Solid-state NMR | / | 2H3O |
| MerF Hg (II) transporter, truncated protein (AAs 13-72) determined in SDS micelles (expressed in E. coli) | Morganella morganii | Solution NMR | / | 1WAZ |
| MerF Hg (II) transporter, truncated protein (AAs 13-70) determined in proteoliposomes (expressed in E. coli) | Morganella morganii | Solid-state NMR | / | 2LJ2 |
Table 1. Structural Research of Bacterial Mercury Detoxification Proteins.
At Creative Biostructure, our accomplished and adept team of veteran scientists and structural biologists possess an impressive amount of extensive experience in the intricacies of analyzing the structural properties of even the most demanding targets, such as the elusive membrane proteins. With great efficacy, we have surmounted the formidable challenge of determining the elusive structures of countless membrane proteins including but not limited to the ion channels, transporters, and receptors. Our scientific enterprise has ceaselessly aided our esteemed clients in gaining previously unattainable insights into the enigmatic mechanisms governing the intricate functions and regulations of proteins. Our structural biology services are fully customizable and entirely bespoke to suit the unique research needs of each of our clients. Upheld by our investment in and utilization of state-of-the-art instrumentation and revolutionary software, allows us to achieve the zenith of perfection and excellence in the domains of data analysis and modeling. For an unparalleled degree of professionalism and an all-encompassing solution that is guaranteed to meet your structural biology needs, do not hesitate to contact us.
References
- De Angelis A A, et al. Structure determination of a membrane protein with two trans-membrane helices in aligned phospholipid bicelles by solid-state NMR spectroscopy. Journal of the American Chemical Society. 2006, 128(37): 12256-12267.
- Howell S C, et al. NMR structure determination of a membrane protein with two transmembrane helices in micelles: MerF of the bacterial mercury detoxification system. Biochemistry. 2005, 44(13): 5196-5206.
- Das B B, et al. Structure determination of a membrane protein in proteoliposomes. Journal of the American Chemical Society. 2012, 134(4): 2047-2056.