Selection for Binding Affinity
Based on the comprehensive protein engineering platform through many years of experience, experts from Creative Biostructure can provide excellent custom selection for binding affinity services in directed evolution. Binding affinity is an important component of enzymatic activities, catalytic efficiency and the rate of product release, which can strongly determine overall enzyme desirability.
Creative Biostructure has strong expertise in well-designed selection at the expense of potentially rich screening data. Selection can bypass the requirement to individually inspect each library membrane and instead links an activity of interest to physical separation of the encoding DNA or to survival of the organism producing active library members. In a typical target-binding selection, protein library members with desired binding activity can be captured using an immobilized target, whereas non-binding library members will be washed away. Notably, in phage display or cell surface display technique, a cell or bacteriophage function as a compartment to link genes and gene products. Thus, protein library members are expressed on the surface of the cell or the coat of the bacteriophage through fusion with endogenous cell surface proteins or phage coat proteins.
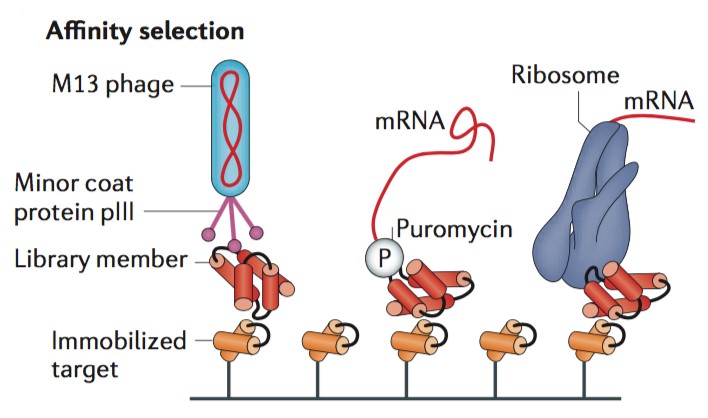 Figure 1. Schematic of affinity selection. (Nature Reviews Genetics, 2015)
Figure 1. Schematic of affinity selection. (Nature Reviews Genetics, 2015)
Creative Biostructure can perform binding affinity selection services through cell surface or phage display methods that require intracellular translation, which holds the library size limitations during screening methods. It has been reported that bacterial transformation provides around 109-1010 transformants per experiment, while cell- or phage-based selection methods are generally limited to library sizes in this range. Therefore, Creative Biostructure provides an alternative method, ribosome display, can bypass this bottleneck through the utility of in vitro translation reactions. In the absence of the stop codon and under controlled conditions, ribosomes remain stably bound to both mRNA and the growing polypeptide, thereby coupling proteins with their encoding genes. This technique developed by Creative Biostructure can also be applied for selection of antibodies based upon their binding strength.
Creative Biostructure’s custom screening or selection services can provide you a wide range of first-class strategies for directed evolution. Creative Biostructure also provides Membrane Protein Platform and Phage Display Platform for your specific projects. Please feel free to contact us for a detailed quote.
References:
M. S. Packer and D. R. Liu (2015). Methods for the directed evolution of proteins. Nature Reviews Genetics, 16: 379-394.
R. E. Hawkins, et al. (1992). Selection of phage antibodies by binding affinity. Mimicking affinity maturation. J. Mol. Biol., 226(3): 889-896.
Selection and amplification binding assay. (https://en.wikipedia.org/wiki/Selection_and_amplification_binding_assay)