Structural Research of Superfamily of K+ Transporters (SKT proteins)
Members of the superfamily of K+ transporter proteins (SKT) are integral membrane proteins that mediate the uptake of ions in non-animal cells and play an essential role in ion homeostasis. Research has found that SKT proteins are homologous to tetrameric K+ channels and mediate the rapid and selective flow of K+ along their electrochemical gradients.
Structural elucidation of the K+ transport complex of prokaryotes
KdpFABC is an oligomeric K+ transport complex in prokaryotes that maintains ionic homeostasis under adverse conditions and regulates pH, membrane potential, and swelling pressures that drive cell growth and division. The complex includes a channel-like subunit (KdpA) from the K+ transporter protein superfamily and a pump-like subunit (KdpB) from the p-type atpase superfamily. The corresponding structures were determined by cryo-electron microscopy, and analyses of the sequence and topology of KdpA confirmed that it contains four nearly repetitive "MPM" folds, which are very similar to the bacterial potassium channels TrkH and KtrB.
Action mechanism of the SKT protein KdpA in K+ transport
In KdpA, K+ ions are bound within a selective filter (SF), and, like TrkH and KtrB, the third MPM repeat sequence has a kinked helix with a loop that prevents ions from crossing the membrane. This loop acts as a gate that can be moved aside to open a pore for transport. In addition, the researchers found a 40 Å-long tunnel encapsulated within the membrane domain that connects the SF in KdpA to the Bx site in KdpB for charge transfer between these sites. the binding of K+ to the SF triggers the passage of protons through the tunnel.
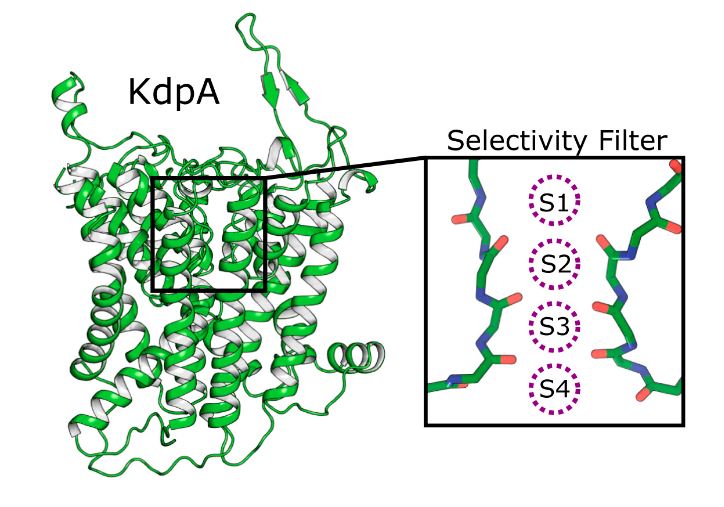 Figure 1. Four binding sites for K+ in SKT protein KdpA. (Sweet ME, et al., 2021)
Figure 1. Four binding sites for K+ in SKT protein KdpA. (Sweet ME, et al., 2021)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Square-shaped octameric of KtrA with ATP bound | Bacillus subtilis subsp. subtilis str. 168 | X-ray diffraction | 3.24 Å | 4J90 |
| Square-shaped octameric of KtrA with ADP bound | Bacillus subtilis subsp. subtilis str. 168 | X-ray diffraction | 2.93 Å | 4J91 |
| Potassium transporter TrkH | Vibrio parahaemolyticus | X-ray diffraction | 3.506 Å | 3PJZ |
| TrkH-TrkA in complex with ATP | Vibrio parahaemolyticus RIMD 2210633 | Cryo-EM single particle analysis | 2.97 Å | 6V4J |
| TrkH-TrkA in complex with ADP | Vibrio parahaemolyticus | X-ray diffraction | 3.53 Å | 6V4K |
| TrkH-TrkA in complex with ATPgammaS | Vibrio parahaemolyticus RIMD 2210633 | X-ray diffraction | 3.8 Å | 6V4L |
| TrkH/TrkA potassium transport complex | Vibrio parahaemolyticus RIMD 2210633 | X-ray diffraction | 3.8 Å | 4J9U |
| KtrAB potassium transporter | Bacillus subtilis subsp. subtilis str. 168 | X-ray diffraction | 3.5 Å | 4J7C |
| E1-ATP KdpFABC complex. | Escherichia coli | Cryo-EM single particle analysis | 3.2 Å | 7NNP |
| KdpFABC complex | Escherichia coli | X-ray diffraction | 2.9 Å | 5MRW |
| KdpFABC in E1 state with K | Escherichia coli K-12 | Cryo-EM single particle analysis | 3.38 Å | 7BH1 |
| KdpFABC in E1-ATP state | Escherichia coli K-12 | Cryo-EM single particle analysis | 3.23 Å | 7LC3 |
| KdpFABC in E2-P state with BeF3 | Escherichia coli K-12 | Cryo-EM single particle analysis | 3.7 Å | 7LC6 |
| KdpFABC in E2Pi state with MgF4 | Escherichia coli K-12 | Cryo-EM single particle analysis | 2.9 Å | 7BGY |
| KdpFABC in E2Pi state with BeF3 and K+ | Escherichia coli K-12 | Cryo-EM single particle analysis | 3 Å | 7BH2 |
| KdpFABC complex in an E1-ATP conformation loaded with K+ | Escherichia coli | Cryo-EM single particle analysis | 3.1 Å | 7NNL |
| KdpFABC complex in the E2-P conformation, under turnover conditions | Escherichia coli K-12 | Cryo-EM single particle analysis | 4 Å | 7ZRL |
| KdpFABC complex in an E1 outward-facing state (state 1) | Escherichia coli K-12 | Cryo-EM single particle analysis | 3.7 Å | 6HRA |
| KdpFABC complex in an E2 inward-facing state (state 2) | Escherichia coli K-12 | Cryo-EM single particle analysis | 4 Å | 6HRB |
| KdpFABC complex in a nucleotide-free E1 conformation loaded with K+ | Escherichia coli | Cryo-EM single particle analysis | 3.4 Å | 7ZRH |
| KdpFABC complex in the E1-P_ADP conformation, under turnover conditions | Escherichia coli | Cryo-EM single particle analysis | 3.7 Å | 7ZRM |
| KdpFABC complex in a nucleotide-free E1 conformation loaded with K+ | Escherichia coli | Cryo-EM single particle analysis | 3.5 Å | 7ZRI |
| KdpFABC complex in a nucleotide-free E1 conformation loaded with K+ | Escherichia coli | Cryo-EM single particle analysis | 3.7 Å | 7ZRJ |
Table 1. Structural research of the superfamily of K+ transporters (SKT proteins).
Creative Biostructure can offer a full suite of cutting-edge protein structural analysis services. Our advanced technologies include X-ray crystallography, cryo-electron microscopy (cryo-EM), and NMR spectroscopy, which allow us to delve deeper into the three-dimensional structure of proteins and provide valuable insights into their function and regulation. In addition to our commitment to high-quality results, we offer competitive pricing and flexible service options to satisfy the diverse requirements of our clients.
Our experienced scientists are ready to address any questions or concerns. If you would like to understand the complex structure of proteins and other biomolecules, please contact us for a professional and comprehensive solution.
References
- Sweet ME, et al. Structural basis for potassium transport in prokaryotes by KdpFABC. Proc Natl Acad Sci U S A. 2021. 118(29): e2105195118.
- Levin EJ, Zhou M. Recent progress on the structure and function of the TrkH/KtrB ion channel. Curr Opin Struct Biol. 2014. 27: 95-101.