Structural Research of Stomatin, Prohibitin, Flotillin, and HflK/C (SPFH) Family Proteins
The stomatin, phibitin, flotillin, and hflk/c (SPFH) family proteins are enriched in functional membrane microstructural domains (FMMs) at various subcellular locations and have been hypothesized to act as scaffolds during microstructural domain formation. In addition, many SPFH proteins are associated with highly specific processes occurring in membranes. In recent years, researchers have used cutting-edge imaging techniques to understand the function of SPFH proteins at the molecular level.
Structural analysis of two bacterial SPFH proteins
Researchers report the structures of supramolecular complex SPFH proteins (HflK and HflC) isolated from bacterial membrane microdomains. The crystal structures show that HflK and HflC form a cyclic 24-polymer aggregate with laterally separated membrane microdomains 20 nm in diameter, bounded by transmembrane domains and a completely sealed periplasmic dome. Four membrane-anchored AAA+ protease (FtsH) hexamers are embedded in this microdomain through interaction with the inner surface of the dome. These observations provide a mechanistic explanation for the action mechanism by which HflK/C and its mitochondrial homolog, inhibin, regulate membrane-bound AAA+ protease.
Preparation of the SPFH complexes
Overexpressed SPFH protein is purified from E. coli BL21 (DE3) cell lysate membrane fractions. The purification buffer is tested and evaluated by negative staining electron microscopy (nsEM) in detergent and different concentrations of glycerol used for the preparation of the complexes. To further improve sample homogeneity, glycerol density gradient centrifugation is used and peak fractions are used for functional and structural characterization. Each fraction is examined using nsEM. The complete complexes are then assayed for protein degradation using the reported substrates.
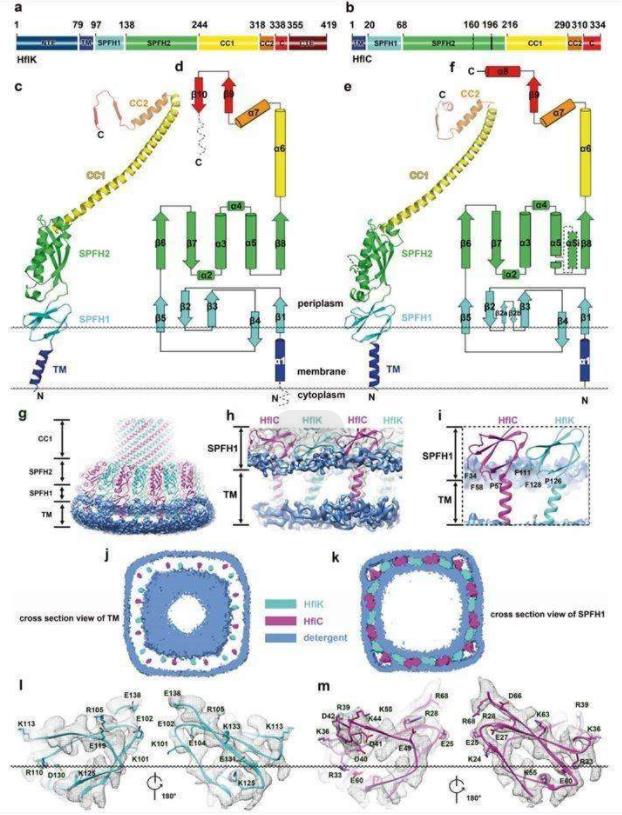 Figure 1. Structure of HflK and HflC. (Ma C, et al., 2022)
Figure 1. Structure of HflK and HflC. (Ma C, et al., 2022)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| SPFH domain of stomatin | Pyrococcus horikoshii | X-ray diffraction | 2.5 Å | 8GN9 |
| SPFH domain of stomatin (Crystal form 1) | Mus musculus | X-ray diffraction | 2.46 Å | 4FVF |
| SPFH domain of stomatin (Crystal form 2) | Mus musculus | X-ray diffraction | 2.69 Å | 4FVJ |
| SPFH domain of stomatin (Crystal form 3) | Mus musculus | X-ray diffraction | 1.8 Å | 4FVG |
| A core domain of stomatin | Pyrococcus horikoshii | X-ray diffraction | 3.2 Å | 3BK6 |
| SPFH family proteins | Escherichia coli | Cryo-EM single particle analysis | 3.27 Å | 7VHQ |
| SPFH family proteins | Escherichia coli | Cryo-EM single particle analysis | 3.27 Å | 7VHP |
| FtsH-HflkC AAA protease complex | Escherichia coli K-12 | Cryo-EM single particle analysis | 4 Å | 7WI3 |
| C-terminal domain of stomatin operon partner protein 1510-C | Pyrococcus horikoshii OT3 | X-ray diffraction | 2.4 Å | 3WWV |
| Stomatin SPFH domain in a phosphate buffer | Homo sapiens | SOLUTION NMR | / | 7WH3 |
| Band 7 domain of the flotillin 2 protein | Mus musculus | SOLUTION NMR | / | 1WIN |
| Prohibitin 2 | Homo sapiens | X-ray diffraction | 1.701 Å | 6IQE |
Table 1. Structural research of the stomatin, prohibitin, flotillin, and hflk/c (SPFH) family proteins.
Creative Biostructure is a biotechnology company specializing in protein structural biology and provides structure analysis services for SPFH family proteins. We have cutting-edge equipment and technology to execute a variety of structural analysis methods, including X-ray crystallography, cryo-electron microscopy (cryo-EM), and NMR spectroscopy.
Through our advanced services, we can help you perform groundbreaking protein structure research to further resolve the higher-order formation and molecular mechanisms. If you are interested in our services, please contact us for more details.
References
- Ma C, et al. Structural insights into the membrane microdomain organization by SPFH family proteins. Cell Research. 2022. 32(2): 176-189.
- Scholz AS, et al. An Stomatin, Prohibitin, Flotillin, and HflK/C-Domain Protein Required to Link the Phage-Shock Protein to the Membrane in Bacillus subtilis. Front Microbiol. 2021. 12: 754924.
- Yokoyama H, Matsui I. Higher-order structure formation using refined monomer structures of lipid raft markers, Stomatin, Prohibitin, Flotillin, and HflK/C-related proteins. FEBS Open Bio. 2023. 13(5): 926-937.