Structural Research of Claudins
Claudins play an important role in the formation and maintenance of tight junctions (TJs) in epithelial and endothelial cells. TJs are specialized structures that connect adjacent cells and create a barrier that regulates the movement of molecules and ions across the epithelium. Clostridium perfringens is a human pathogenic bacterium that is known to secrete a virulent enterotoxin called CpE. This toxin exhibits a deleterious effect on the gut epithelial barrier by targeting claudins. The C-terminal receptor-binding domain (cCpE) of CpE binds to claudins in an isoform-specific manner, causing the dissociation of TJs and leading to cytotoxicity.
One of the most significant breakthroughs in the structural research of claudins has been the determination of their crystal structure. In a recent study, researchers used X-ray crystallography to determine the crystal structure of human claudin-9 (hCLDN-9) in a complex with cCpE, which revealed the mechanism for toxin-induced gut barrier breakdown. The study found that hCLDN-9 is a high-affinity receptor for CpE and that cCpE induces alterations to two epitopes that are known to enable claudin self-assembly, resulting in exposure of high-affinity interactions between hCLDN-9 and cCpE. This helps in understanding isoform-specific recognition of claudins by CpE.
The crystal structure of Claudin-9 provides valuable insights into the structure and function of other Claudin family members. It also highlights the importance of structural research in understanding the molecular mechanisms underlying tight junction formation and regulation.
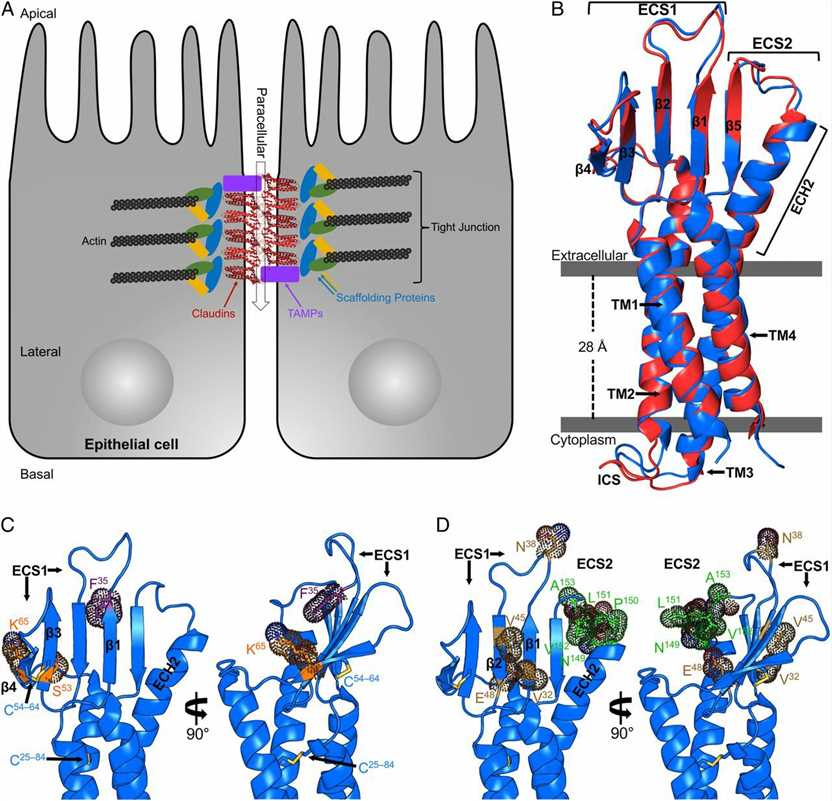 Figure 1. Structures of TJs and hCLDN-9. (Vecchio A J, et al., 2019)
Figure 1. Structures of TJs and hCLDN-9. (Vecchio A J, et al., 2019)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Claudin-4 in complex with Clostridium perfringens enterotoxin C-terminal domain (expressed in Trichoplusia ni) | Homo sapiens | X-ray diffraction | 3.37 Å | 7KP4 |
| Claudin-4 in complex with Clostridium perfringens enterotoxin C-terminal domain in complex with sFab COP-2 (expressed in Trichoplusia ni) | Homo sapiens | Cryo-EM single particle analysis | 6.90 Å | 7TDM |
| Claudin-9 in complex with Clostridium perfringens enterotoxin C-terminal domain, closed form (expressed in Trichoplusia ni) | Homo sapiens | X-ray diffraction | 3.20 Å | 6OV2 |
| Caludin-15 (expressed in S. frugiperda) | Mus musculus | X-ray diffraction | 2.40 Å | 4P79 |
| Claudin-19 in complex with Clostridium perfringens enterotoxin (expressed in S. frugiperda) | Mus musculus | X-ray diffraction | 3.70 Å | 3X29 |
Table 1. Structural Research of Claudins.
Creative Biostructure offers a wide range of services to support the structural research of claudins and other membrane proteins. Our state-of-the-art facilities and experienced scientists are capable of providing high-quality membrane protein production, crystallization, and structure determination services.
Our membrane protein production service includes gene cloning, protein expression, and purification, which can be customized according to specific client requirements. Our membrane protein crystallization service utilizes a variety of techniques, including vapor diffusion, microbatch, and lipidic cubic phase (LCP), to optimize protein crystallization conditions and maximize the success rate of structure determination. Our structure determination service includes X-ray crystallography, NMR spectroscopy, and cryo-electron microscopy (cryo-EM), which are widely used techniques in the structural research of membrane proteins. We also provide advanced data processing and analysis services to ensure high-quality structural data and accurate interpretation of the results. If you are interested in learning more about our protein structure analysis services, please contact us for more information.
References
- Vecchio A J, Stroud R M. Claudin-9 structures reveal mechanism for toxin-induced gut barrier breakdown. Proceedings of the National Academy of Sciences. 2019, 116(36): 17817-17824.
- Vecchio A J, Rathnayake S S, Stroud R M. Structural basis for Clostridium perfringens enterotoxin targeting of claudins at tight junctions in mammalian gut. Proceedings of the National Academy of Sciences. 2021, 118(15): e2024651118.