Structural Research of Phospholipid Phosphatases
Phosphatidic acid phosphatase (PAP) catalyzes the dephosphorylation of phosphatidic acid to produce diacylglycerols and inorganic phosphates. In eukaryotic cells, PAP activity plays a central role in the synthesis of phospholipids and triglycerides through its product, glycerol diesters. It also produces and/or degrades lipid signaling molecules associated with phosphatidic acid. Mutations in PAP inactivation result in rhabdomyolysis, autoinflammatory diseases, and abnormal fat storage. Lipin/Pah PAP regulates energy storage, energy mobilization, lipogenesis, phospholipid synthesis, autophagy, celiac secretion, and fatty acid synthesis.
There are two types of PAP enzymes, Mg2+-dependent (PAP1) and Mg2+-independent (PAP2), and studies have identified the gene encoding the PAP2 enzyme in Saccharomyces cerevisiae brewer's yeast (PAH1). This discovery reveals the molecular function of mammalian lipoproteins, whose deficiency leads to impaired lipid metabolism in mice. PAPs are a key class of membrane proteins that play a crucial role in cellular lipid metabolism and signaling.
The structure of lipid/Pah PAPs varies, but all PAPs retain two regions called N-Lip and C-Lip, originally named for their location at the opposite ends of human and mouse lipins 4. C-Lip contains the catalytic structural domain from the haloalkanoic acid dehalogenase (HAD) superfamily 20 and has the catalytic DxDxT motif. PAP activity requires N-Lip, but the functional prediction of N-Lip is complicated by the lack of sequence homology with known functional domains and the fact that N-Lip is distinct to PAP. N-Lip is separated from C-Lip by an extended splice that typically contains 250-500 residues. The splice sequence is highly variable but retains conserved functions that regulate lipin/Pah-PAP localization and activity.
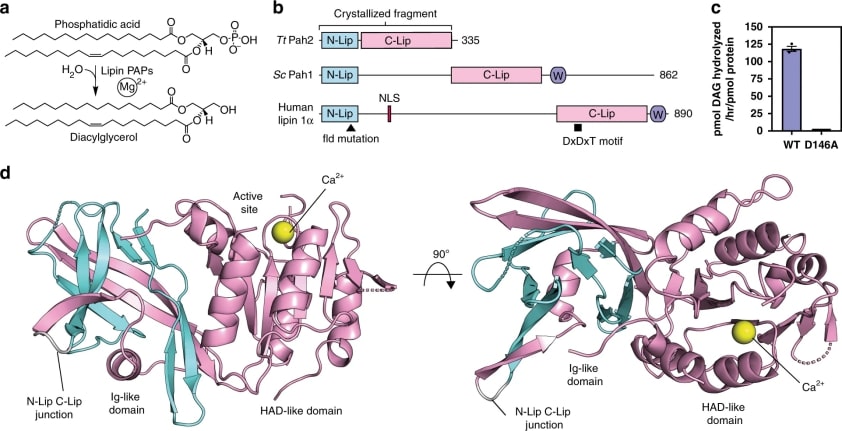 Figure 1. Structure of TtPah2 reveals the N-Lip co-folds with the C-Lip to form a split immunoglobulin-like domain. (Khayyo
VI, et al., 2020)
Figure 1. Structure of TtPah2 reveals the N-Lip co-folds with the C-Lip to form a split immunoglobulin-like domain. (Khayyo
VI, et al., 2020)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| MTMR8 catalytic phosphatase domain: Homo sapiens E (expressed in E. coli) | Homo sapiens | X-ray diffraction | 2.8 Å | 4Y71 |
| lipin/Pah phosphatidic acid phosphatase: Tetrahymena thermophila B (expressed in E. coli) | Tetrahymena thermophila | X-ray diffraction | 3 Å | 6TZY |
Table 1. Structural Research of Phospholipid Phosphatases.
The Significance of Studying the Structure of Phospholipid Phosphatases
- The structure of phospholipid phosphatase proteins can explain their catalytic mechanism and substrate specificity.
- Phospholipid phosphatases play a vital role in cellular lipid metabolism, signal transduction, and disease pathogenesis. Their structural resolution can help reveal their biological functions and potential drug targets.
- Structural studies of phospholipid phosphatase proteins provide significant information and a basis for drug design and development.
In recent years, several significant breakthroughs and discoveries have been made by using various structural biology methods and techniques, including X-ray crystallography, electron microscopy, and nuclear magnetic resonance, to study phospholipid phosphatases. These studies have revealed the relationship between the structure and function of phospholipid phosphatases.
Creative Biostructure is a scientific research and services company focused on protein structural biology. We offer a wide range of technologies and services including X-ray crystallography, cryo-electron microscopy (Cryo-EM), and NMR services. Our team of professionals has the experience and expertise to provide a full range of support and solutions for your research. Whether it is the crystal structure, electron microscope structure, or NMR structure, we will provide you with high-quality data and analysis results according to your needs and research objectives to help you gain insight into the structure and function of phospholipid phosphatase proteins and advance your research to breakthroughs. If you are interested in our services, please contact us for a more detailed description of our services.
References
- Carman GM,Han GS. Roles of phosphatidate phosphatase enzymes in lipid metabolism. Trends Biochem Sci. 2006; 31 (12): 694-9.
- Khayyo VI, et al. Crystal structure of a lipin/Pah phosphatidic acid phosphatase. Nat Commun. 2020; 11 (1): 1309.