Structural Research of Cys-Loop Receptor Family
Ligand-gated ion channels (LGICs) of the Cys-loop receptor family are composed of nicotinic acetylcholine, GABAA, GABAA-ρ, glycine, 5-HT3 receptors, and zinc-activated (ZAC) receptors. These ion channels are involved in neurotransmission and are targets for many drugs, including anesthetics, sedatives, and muscle relaxants. These receptors are pentameric, meaning they consist of five protein subunits that form a pentameric arrangement around a central pore.
In recent years, there has been significant progress in the structural research of the Cys-loop receptor family. Researchers have utilized a variety of techniques, including X-ray crystallography, cryo-electron microscopy (cryo-EM), and molecular dynamics simulations, to better understand the receptor's structure and function. X-ray crystallography has been used to determine the structures of acetylcholine receptor and the glycine receptor. These structures have provided insights into the architecture of the receptor, including the arrangement of subunits and the location of key amino acid residues involved in ligand binding and channel gating. The cryo-EM technique has enabled researchers to study the receptor in different conformations and to visualize the dynamics of the receptor's movement during ion channel gating. Molecular dynamics simulations have been used to study the behavior of these proteins in lipid bilayers and to provide insights into the receptor's interaction with its environment. These simulations have also been used to predict the effect of mutations on the receptor's function and to design novel ligands that can target specific regions of the receptor.
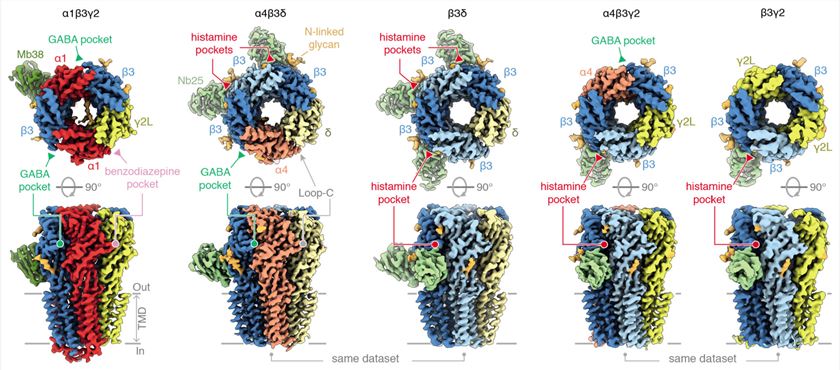 Figure 1. Landscape of differential GABAA receptor assemblies. (Sente A, et al., 2022)
Figure 1. Landscape of differential GABAA receptor assemblies. (Sente A, et al., 2022)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Nicotinic Acetylcholine Receptor Pore (closed state) | Torpedo marmorata | Electron diffraction | 4.00 Å | 1OED |
| Nicotinic Acetylcholine Receptor, refined structure | Torpedo marmorata | Electron diffraction | 4.00 Å | 2BG9 |
| Acetylcholine receptor analyzed by time-resolved electron cryo-microscopy (closed class) | Torpedo marmorata | Time-resolved electron cryo-microscopy | 6.20 Å | 4AQ5 |
| Acetylcholine (ACh) receptor in complex with alpha-bungarotoxin in nanodiscs | Tetronarce californica | Cryo-EM single particle analysis | 2.69 Å | 6UWZ |
| Acetylcholine (ACh) receptor, apo form | Tetronarce californica | Cryo-EM single particle analysis | 2.50 Å | 7SMM |
| Nicotinic acetylcholine (ACh) receptor, resting conformation | Tetronarce californica | Cryo-EM single particle analysis | 2.90 Å | 7QKO |
| Acetylcholine (ACh) receptor in complex with alpha-neurotoxin in nanodiscs (expressed in E. coli) | Tetronarce californica | Cryo-EM single particle analysis | 3.15 Å | 7Z14 |
| Nicotinic Acetylcholine α4β2 Receptor (expressed in HEK293S) | Homo sapiens | X-ray diffraction | 3.94 Å | 5KXI |
| Nicotinic Acetylcholine α4β2 Receptor, 2α3β stiochiometry (expressed in HEK cells) | Homo sapiens | Cryo-EM single particle analysis | 3.70 Å | 6CNJ |
| Nicotinic Acetylcholine α3β4 Receptor (expressed in HEK293 cells) | Homo sapiens | Cryo-EM single particle analysis | 3.34 Å | 6PV7 |
| Nicotinic Acetylcholine α4β2 Receptor with varenicline in complex with anti-BRIL synthetic antibody BAK5 (expressed in HEK293 cells) | Homo sapiens | Cryo-EM single particle analysis | 3.87 Å | 6USF |
| Prokaryotic pentameric ligand-gated ion channel (ELIC) (expressed in E. coli) | Dickeya chrysanthemi | X-ray diffraction | 3.30 Å | 2VL0 |
| Prokaryotic pentameric ligand-gated ion channel (ELIC) in complex with acetylcholine (expressed in E. coli) | Dickeya dadantii | X-ray diffraction | 2.91 Å | 3RQW |
| Prokaryotic pentameric ligand-gated ion channel (ELIC) in complex with bromoform (expressed in E. coli) | Dickeya chrysanthemi | X-ray diffraction | 3.65 Å | 3ZKR |
| Prokaryotic pentameric ligand-gated ion channel (ELIC) in complex with Br-memantine (expressed in E. coli) | Dickeya chrysanthemi | X-ray diffraction | 3.20 Å | 4TWD |
| Prokaryotic pentameric ligand-gated ion channel, 7'C pore mutant (L238C) (expressed in E. coli) | Dickeya chrysanthemi | X-ray diffraction | 2.50 Å | 6HJX |
| Prokaryotic pentameric ligand-gated ion channel (ELIC) in POPC-only nanodiscs (expressed in E. coli) | Dickeya dadantii | Cryo-EM single particle analysis | 4.10 Å | 6V0B |
| Prokaryotic pentameric ligand-gated ion channel (ELIC) with PAM nanobody (expressed in E. coli) | Dickeya chrysanthemi | X-ray diffraction | 2.59 Å | 6SSI |
| Prokaryotic pentameric ligand-gated ion channel (GLIC) (expressed in E. coli) | Gloeobacter violaceus | X-ray diffraction | 3.10 Å | 3EHZ |
| Prokaryotic pentameric ligand-gated ion channel (GLIC) (expressed in E. coli) | Gloeobacter violaceus | X-ray diffraction | 2.90 Å | 3EAM |
| Prokaryotic pentameric ligand-gated ion channel (GLIC), wildtype-TBSb complex (expressed in E. coli) | Gloeobacter violaceus | X-ray diffraction | 3.70 Å | 2XQA |
| Prokaryotic pentameric ligand-gated ion channel (GLIC) (expressed in E. coli) | Gloeobacter violaceus | X-ray diffraction | 2.40 Å | 4HFI |
| Prokaryotic "pentameric" ligand-gated ion channel (GLIC) with hexameric quaternary structure (expressed in E. coli) | Gloeobacter violaceus | X-ray diffraction | 2.30 Å | 3IGQ |
| Prokaryotic pentameric ligand-gated ion channel (GLIC) in complex with propofol anesthetic (expressed in E. coli) | Gloeobacter violaceus | X-ray diffraction | 3.30 Å | 3P50 |
| Prokaryotic pentameric ligand-gated ion channel (GLIC) in complex with ketamine anesthetic (expressed in E. coli) | Gloeobacter violaceus | X-ray diffraction | 2.99 Å | 4F8H |
| Prokaryotic pentameric ligand-gated ion channel (GLIC), pH 4 (expressed in Drosophila melanogaster) | Gloeobacter violaceus | X-ray diffraction | 3.35 Å | 4NPP |
| Prokaryotic pentameric ligand-gated ion channel (GLIC) (expressed in E. coli) | Gloeobacter violaceus | X-ray diffraction | 3.00 Å | 4QH5 |
| Prokaryotic pentameric ligand-gated ion channel (GLIC), Bromoform bound, K33C-L246C mutant (expressed in E. coli) | Gloeobacter violaceus | X-ray diffraction | 2.95 Å | 5HCJ |
| Prokaryotic pentameric ligand-gated ion channel (GLIC) with bound thiopental (expressed in E. coli) | Gloeobacter violaceus | X-ray diffraction | 3.50 Å | 5L4E |
| Prokaryotic pentameric ligand-gated ion channel (GLIC) in complex with DHA (expressed in E. coli) | Gloeobacter violaceus | X-ray diffraction | 3.25 Å | 5J0Z |
| Prokaryotic pentameric ligand-gated ion channel (GLIC), open channel-stabilized mutant G-2'I + I9'A (expressed in E. coli) | Gloeobacter violaceus | X-ray diffraction | 3.12 Å | 5V6O |
| Prokaryotic pentameric ligand-gated ion channel (GLIC), H235Q apo mutant (expressed in E. coli) | Gloeobacter violaceus | X-ray diffraction | 2.95 Å | 5NJY |
| Pentameric ligand-gated ion channel (GLIC), wild-type (expressed in E. coli) | Gloeobacter violaceus | X-ray diffraction | 2.22 Å | 6HZW |
| Prokaryotic pentameric ligand-gated ion channel (GLIC), pH 7 (expressed in E. coli) | Gloeobacter violaceus | Cryo-EM single particle analysis | 4.10 Å | 6ZGD |
| Prokaryotic pentameric ligand-gated ion channel (ELIC) in POPC nanodisc (expressed in E. coli) | Dickeya dadantii | Cryo-EM single particle analysis | 3.14 Å | 8D63 |
| Prokaryotic pentameric ligand-gated ion channel (pLGIC) with additional N-terminal domain, closed pore (expressed in E. coli) | Desulfofustis | X-ray diffraction | 3.55 Å | 6V4S |
| Prokaryotic pentameric ligand-gated ion channel (sTeLIC), wild-type (expressed in E. coli) | endosymbiont of Tevnia jerichonana | X-ray diffraction | 2.30 Å | 6FL9 |
| Human glycine receptor (hGlyR-α1) transmembrane domain monomer (expressed in E. coli) | Homo sapiens | Solution NMR | / | 2M6B |
| Human glycine receptor (hGlyR-α3) in complex with strychnine (expressed in Spodoptera frugiperda) | Homo sapiens | X-ray diffraction | 3.04 Å | 5CFB |
| Human glycine receptor (hGlyR-α3) N38Q mutant in complex with AM-3607 (expressed in Spodoptera frugiperda) | Homo sapiens | X-ray diffraction | 2.61 Å | 5TIN |
| Human glycine receptor (hGlyR-α3) in complex with Gly and ivermectin (expressed in Spodoptera frugiperda) | Homo sapiens | X-ray diffraction | 2.85 Å | 5VDH |
| alpha-1 GlyR Glycine receptor in complex with strychnine (expressed in Spodoptera frugiperda) | Danio rerio | Cryo-EM single particle analysis | 3.90 Å | 3JAD |
| alpha-1 GlyR Glycine receptor, full-length in nanodiscs. Apo/Resting conformation (expressed in Spodoptera frugiperda) | Danio rerio | Cryo-EM single particle analysis | 3.33 Å | 6UBS |
| alpha-1 glycine receptor bound with glycine in nanodisc, desensitized state (expressed in Spodoptera frugiperda) | Danio rerio | Cryo-EM single particle analysis | 3.20 Å | 6PLR |
| Full length alpha1 Glycine receptor in presence of 32uM Tetrahydrocannabinol (expressed in Spodoptera frugiperda) | Danio rerio | Cryo-EM single particle analysis | 3.09 Å | 7M6M |
| Glutamate-gated chloride channel (GluCl) in complex with Fab and ivermectin (expressed in Spodoptera frugiperda) | Caenorhabditis elegans | X-ray diffraction | 3.26 Å | 3RHW |
| alpha7 nAChR transmembrane domain (expressed in E. coli) | Homo sapiens | Solution NMR | / | 2MAW |
| alpha-7 nicotinic acetylcholine receptor bound to alpha-bungarotoxin in a resting state (expressed in HEK293 cells) | Homo sapiens | Cryo-EM single particle analysis | 3.00 Å | 7KOO |
| alpha 7 nicotinic acetylcholine receptor in apo-form (expressed in HEK293 cells) | Homo sapiens | Cryo-EM single particle analysis | 3.18 Å | 7EKI |
| alpha7 neuronal ACh receptor, intracellular domain with transmembrane domain (expressed in E. coli) | Homo sapiens | Solution NMR | / | 7RPM |
| GABAA receptor (β3 homopentamer) (expressed in HEK293F cells) | Homo sapiens | X-ray diffraction | 2.97 Å | 4COF |
| GABAA receptor (α1β2γ2) in complex with GABA and flumazenil antagonist (expressed in HEK293 cells) | Homo sapiens | Cryo-EM single particle analysis | 3.92 Å | 6D6U |
| GABAA receptor (α1β2γ2) in complex with bicuculline methbromide (expressed in HEK293 cells) | Homo sapiens | Cryo-EM single particle analysis | 3.12 Å | 6X3S |
| GABAA receptor (α1β2γ2) in complex with zolpidem (expressed in HEK293 cells) | Homo sapiens | Cryo-EM single particle analysis | 2.90 Å | 8DD2 |
| GABAA receptor (α1β3γ2L) in nanodiscs (expressed in HEK293 cells) | Homo sapiens | Cryo-EM single particle analysis | 3.20 Å | 6I53 |
| GABAA receptor (α1β3γ2L) in complex with picrotoxin (expressed in HEK293S cells) | Homo sapiens | Cryo-EM single particle analysis | 3.10 Å | 6HUG |
| GABAA receptor (β3 homopentamer) in complex with histamine and megabody Mb25 in lipid nanodisc (expressed in HEK293 cells) | Homo sapiens | Cryo-EM single particle analysis | 1.70 Å | 7A5V |
| GABAA receptor (β3 homopentamer) in complex with histamine and megabody Mb25 (expressed in HEK293 cells) | Homo sapiens | Cryo-EM single particle analysis | 2.49 Å | 6QFA |
| GABAA receptor (αββαβ) with bound α-Cobratoxin (expressed in HEK293 cells) | Homo sapiens | Cryo-EM single particle analysis | 3.00 Å | 7PC0 |
| GABAA receptor (α4β3δ), apo (expressed in HEK293 cells) | Homo sapiens | Cryo-EM single particle analysis | 2.50 Å | 7QN5 |
| GABAA receptor in complex with autoimmune antibody Fab175 (expressed in HEK293 cells) | Homo sapiens | Cryo-EM single particle analysis | 3.00 Å | 7T0Z |
| Benzodiazepine-sensitive α1β1γ2S tri-heteromeric GABAA receptor (expressed in TSA201 cells) | Rattus norvegicus | Cryo-EM single particle analysis | 3.80 Å | 6DW0 |
| Serotonin 5-HT3A receptor (expressed in HEK293F cells) | Mus musculus | X-ray diffraction | 3.50 Å | 4PIR |
| Serotonin 5-HT3A receptor (expressed in Spodoptera frugiperda) | Mus musculus | Cryo-EM single particle analysis | 4.31 Å | 6BE1 |
| Serotonin 5-HT3A receptor with bound serotonin, F conformation (expressed in HEK293 cells) | Mus musculus | Cryo-EM single particle analysis | 4.10 Å | 6HIN |
| Serotonin 5-HT3A receptor with bound serotonin, State 1 (expressed in Spodoptera frugiperda) | Mus musculus | Cryo-EM single particle analysis | 3.32 Å | 6DG7 |
| Serotonin 5-HT3A receptor with bound granisetron (expressed in Spodoptera frugiperda) | Mus musculus | Cryo-EM single particle analysis | 2.92 Å | 6NP0 |
| Serotonin 5-HT3A receptor in complex with palonosetron (expressed in HEK293 cells) | Mus musculus | Cryo-EM single particle analysis | 2.82 Å | 6Y1Z |
| Serotonin 5-HT3A receptor in presence of Palonosetron (expressed in Spodoptera frugiperda) | Mus musculus | Cryo-EM single particle analysis | 3.35 Å | 6W1Y |
| Serotonin 5-HT3A receptor with bound serotonin in Salipro bilayer discs (expressed in HEK293 cells) | Mus musculus | Cryo-EM single particle analysis | 2.80 Å | 6Y5A |
Table 1. Structural Research of Cys-Loop Receptor Family.
At Creative Biostructure, we take great pride in offering a comprehensive suite of cutting-edge protein structural analysis services that can aid in the research of the Cys-loop receptor family. Our X-ray crystallography services can determine the high-resolution structure of the receptor, while our cryo-EM services can provide structures of the receptor in different conformations.
Our team of scientists imbued with an almost unparalleled wealth of experience and a relentless commitment to excellence, is passionately dedicated to delivering outstanding results that surpass even the most exacting of client expectations. We harness the most advanced equipment and techniques to painstakingly ensure that our results are both accurate and highly reliable. With our extensive expertise in the field of protein structural analysis, honed over countless years of unparalleled success in the study of membrane proteins, we stand ready to offer our clients valuable insights into the intricacies of the structure and function of the Cys-loop receptor family, thereby opening a veritable treasure trove of new avenues for innovative and groundbreaking research. Contact us to learn more about our services and how we can assist with your research.
References
- Rahman M M, et al. Structure of the native muscle-type nicotinic receptor and inhibition by snake venom toxins. Neuron. 2020, 106(6): 952-962. e5.
- Zhang Y, et al. Asymmetric opening of the homopentameric 5-HT3A serotonin receptor in lipid bilayers. Nature Communications. 2021, 12(1): 1074.
- Sente A, et al. Differential assembly diversifies GABAA receptor structures and signalling. Nature. 2022, 604(7904): 190-194.