Structural Research of Cell Adhesion Molecules
Cell-to-cell and cell-to-Extra Cellular Matrix (ECM) adhesion is one of the most fundamental biological phenomena, and the molecules that mediate this interaction are called Cell adhesion Molecules (CAMs). CAMs are transmembrane glycoproteins present on the surface of the cell membrane or in the ECM, and they usually function as ligands and receptors. More than 50 types of cell adhesion molecules have been identified and are divided into five major groups:
- Integrin Family (IF)
- Immunoglobulin superfamily (SF)
- Selectin family (SF)
- Cadherin Family (CF)
- CD44 molecules
Cell adhesion molecules are transmembrane glycoproteins with a molecular structure consisting of three parts. The first part is the extracellular region, the N-terminal part of the peptide chain with the glycan chain, responsible for the recognition of the ligand. The second part is the transmembrane region, which mostly spans the membrane once. The third part is the cytoplasmic region, the C-terminal part of the peptide chain, which is generally smaller and either directly connected to the backbone components under the plasma membrane or to intracellular chemical signaling molecules to activate the signal transduction pathway.
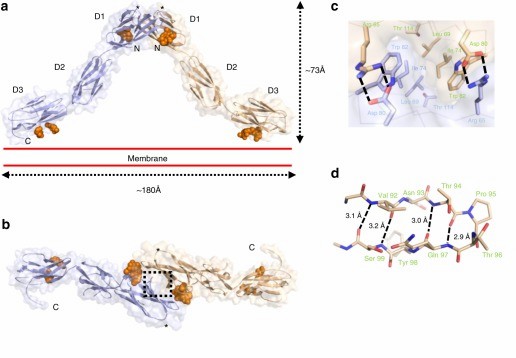 Figure 1. Crystal structure of the OPCML homodimer. (Birtley J R, et al., 2019)
Figure 1. Crystal structure of the OPCML homodimer. (Birtley J R, et al., 2019)
The opioid-like substance binding protein cell adhesion molecule (OPCML) is a glycosylphosphatidylinositol (GPI)-anchored protein whose structure can be revealed by X-ray crystallography. OPCML consists of three Ig-like structural domains, called D1, D2, and D3, connected by short extensional linkers. OPCML homodimers resemble an inverted V shape, mediated by the D1-D1 interface at the distal end of the membrane. The dimerization is stabilized by a combination of hydrophobic contacts, hydrogen bonds, and salt bridges. This interface includes a buried surface area of 874 Å2 and is formed with a solvation-free energy gain (ΔiG) calculated by PISA to be -9.5 kcal M-1. These values are larger than other intermolecular interaction sites observed in crystals, followed by the insertion of the C-terminus of one molecule into the B- and G-chains of D1 in the other molecule. chain and G chain in another molecule, 554 Å2 buried surface area and a solvation-free energy gain of -6.9 kcal M-1. Based on the quaternary structure seen in the asymmetric unit, the dimer will be anchored in the plasma membrane by two D3-linked GPIs. The D3 C-terminus lies approximately 180 Å apart so that the D1 dimer interface at the top of the D1 dimerization interface can extend about 73 Å above the plane of the plasma membrane.
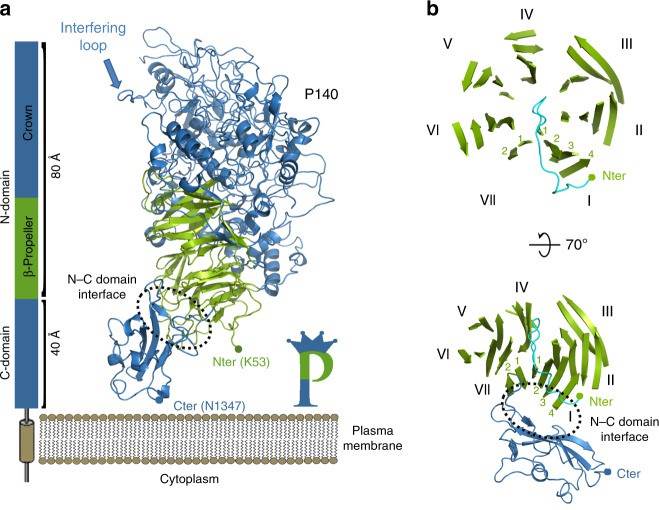 Figure 2. Structure of P140. (Aparicio D, et al., 2020)
Figure 2. Structure of P140. (Aparicio D, et al., 2020)
Essential for reproductive mycoplasma infectivity is a transmembrane adhesion complex called Nap, which contains the proteins P110 and P140. Crystal structures of P140 alone and in complex with the N-terminal structural domain of P110 can be obtained by cryo-EM and tomography (cryo-ET).
The structure of the extracellular region of P140, with a bulky N-terminal structural domain (residues 23-1243) and a small C-terminal structural domain (residues 1244-1351), has an overall shape similar to the capital letter P. The N-terminal structural domain consists of a seven-bladed (β-sheet) propeller and a "crown" formed by the aggregation of long polypeptide segments emerging from the propeller. β-sheets I to VI each have four chains, whereas β-sheet VII, the last β-sheet in the propeller directly connected to the C-terminal structural domain, has only two chains. p140 and p110 share many features in terms of the organization of the structural domain and the topology of the secondary structure elements, although the degree of conservation of the N-terminal and C-terminal structural domains differs.
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Opioid-binding protein cell-adhesion molecule like (OPCML) (expressed in Trichoplusia ni) | Homo sapiens | X-ray diffraction | 2.65 Å | 5UV6 |
| P140-P110 Nap adhesion complex | Mycoplasmoides genitalium G37 | Cryo-EM single particle analysis | 4.10 Å | 6YRK |
| Adhesin P140 | Mycoplasmoides genitalium G37 | X-ray diffraction | 3.24 Å | 6S3U |
| Heterodimer Nap Complex | Mycoplasmoides genitalium G37 | X-ray diffraction | 2.65 Å | 6RUT |
Table 1. Structural Research of Cell Adhesion Molecules.
Cryo-electron microscopy (cryo-EM) is a powerful and rapidly evolving structural biology technique that enables scientists to visualize target membrane proteins at near-atomic resolution. Cryo-EM is also advantageous in its ability to capture the inherent flexibility and dynamics of biomolecules, which is essential for understanding function. By analyzing multiple images of the same sample taken at different angles, a 3D map of the sample can be reconstructed, which can then be used to construct an atomic model of the molecule.
At Creative Biostructure, we can help our customers obtain high-resolution structures of large and complex macromolecular assemblies such as membrane proteins, viruses and ribosomes. If you are interested in learning more about our protein structural analysis services, please contact us for more information.
References
- Birtley J R et al. Inactivating mutations and X-ray crystal structure of the tumor suppressor OPCML reveal cancer-associated functions. Nature Communications, 2019, 10(1).
- Aparicio D, et al. Structure and mechanism of the NAP adhesion complex from the human pathogen Mycoplasma Genitalium. Nature Communications, 2020, 11(1).