Structural Research of Multiple Peptide Resistance Factors (MprFs)
Multiple peptide resistance factors (MprFs) can cause resistance to a variety of pathogens, including antibiotic and cationic antimicrobial peptide resistance. It plays an important role in the metabolism and pathogenicity of bacteria. MprFs utilize two independent domains to synthesize aminoacyl phospholipids and transfer them to the outer lobules of the bacterial membrane. In recent years, research on MprF has found that it can be used as a new antibacterial drug target.
Research Progress on the Mechanism of MprFs
In a study of the relationship between the structure and function of MprF, it was found that bacteria became resistant to CAMP only when the N-terminal domain of MprF caused LES-PG translocation and exposed it to the extracellular lobules. MprF uses L-lysine or L-alanine to modify anionic phospholipids on the membrane surface, thereby introducing a positive charge on the membrane surface. The positive charge introduced reduces its affinity for CAMP.
Research Progress of RtMprFs Structure
The MprF of tropical rhizobia contains a membrane-embedded lipid turnover enzyme domain with two deep cavities facing the inner and outer lobules of the membrane. A hook-like LysPG molecule is located in the inner cavity, with its head group curved toward the outer cavity, which hosts a second phospholipid binding site. In addition, RtMprF exhibits a variety of conformational states, and the synthase domain has a different location relative to the turnover enzyme domain.
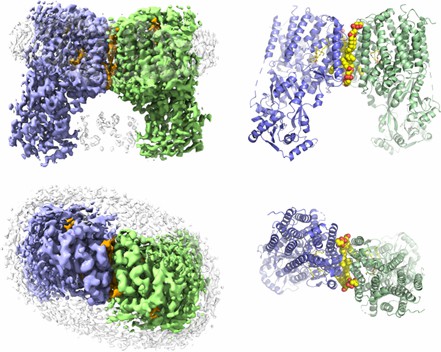 Figure 1. The overall structure of RtMprF(DDM)-nanodic homodimer. (Song D, et al, 2021)
Figure 1. The overall structure of RtMprF(DDM)-nanodic homodimer. (Song D, et al, 2021)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Multiple peptide resistance factor (MprF) loaded with one lysyl-phosphatidylglycerol molecule | Rhizobium tropici CIAT 899 | Cryo-EM single particle analysis | 3.7 Å | 6LVF |
| Multiple peptide resistance factor (MprF) loaded with two lysyl-phosphatidylglycerol molecules | Rhizobium tropici | Cryo-EM single particle analysis | 2.96 Å | 7DUW |
| The soluble domain of the multiple peptide resistance factor (MprF) from Rhizobium tropici | Rhizobium tropici CIAT 899 | X-ray diffraction | 1.998 Å | 6LV0 |
Table 1. Structural research of multiple peptide resistance factors (MprFs).
To study the structure of membrane proteins, we commonly use cryo-electron microscopy (cryo-EM) and X-ray crystallography. Structural analysis of multiple peptide resistance factors (MprFs) is beneficial to the study of the mechanism of MprFs enabling pathogenic bacteria to acquire antimicrobial peptide resistance and the development of MprFS as a target for novel antimicrobial drugs.
Creative Biostructure has long been committed to the study of structural biology and membrane proteins. We have extensive experience in determining membrane protein structures using single particle analysis (SPA).
In addition to the structural determination of membrane proteins, we can accurately analyze biomolecules, including but not limited to nucleic acids, ribosomes, small proteins, protein complexes, protein-ligand complexes, and viruses. If you are interested in our services, please contact us and we will provide you with a professional and comprehensive solution.
References
- Song D, et al. Phospholipid translocation captured in a bifunctional membrane protein MprF. Nat Commun. 2021;12(1):2927.
- Ernst CM, et al. The bacterial defensin resistance protein MprF consists of separable domains for lipid lysinylation and antimicrobial peptide repulsion. PLoS Pathog. 2009;5(11):e1000660.
- Ernst CM, et al. Broad-spectrum antimicrobial peptide resistance by MprF-mediated aminoacylation and flipping of phospholipids. Mol Microbiol. 2011;80(2):290-299.