Structural Research of ADP-Ribosylation Factors
ADP-ribosylation factors (ARFs) are a class of proteins that are widely found in eukaryotic cells. They are small (21 kDa) monomeric GTPases that are important regulators of membrane traffic. When membrane-bound, they recruit soluble junctions to the membrane and trigger the assembly of coating complexes involved in cargo selection and vesicle outgrowth.ARFs, of which there are at least three subclasses and six in total, are small GTP-binding proteins associated with Ras. Their members were initially found to enhance the ADP ribosyltransferase activity of cholera toxin and were subsequently shown to be regulators of intracellular vesicle transport.
The spatial structure of ARFs is also highly conserved with prominent features. n-Myristoylation is a conserved feature of all ARF proteins, which is required for their biological functions.
All ARFs have an a-helix at the N terminus and two identical effector domain regions, Switch 1 and Switch 2, which are connected by two β-fold structures, which we call interwitch. when the N-terminal α-helix is extended, the two amino acid residues in the inter-switch region move, thus enabling the conversion of ARFs from inactive to active conformation, and these two different conformations allow ARF to bind to GDP and GTP, respectively. When GTP binds to ARF, the N-terminal α-helix of ARF protein can insert into the ER membrane, and the GTP-ARF complex binds to the ER membrane, thus initiating the assembly of peripheral proteins.
Solution NMR is a powerful means of analyzing ARFs. For example, the structure of activated (GTP-bound) yeast Arf1 bound to a membrane mimic can be determined by solution NMR methods. Data of primary interest for the experiments include nuclear Overhauser effect (NOE) on fixed-point protonation of fully deuterated preparations, residual dipole coupling (RDC) from 15 N- 1H,15 N- 13C and certain phenyl 13C- 1H bond pairs, and torsional constraints on backbone chemical shift data.
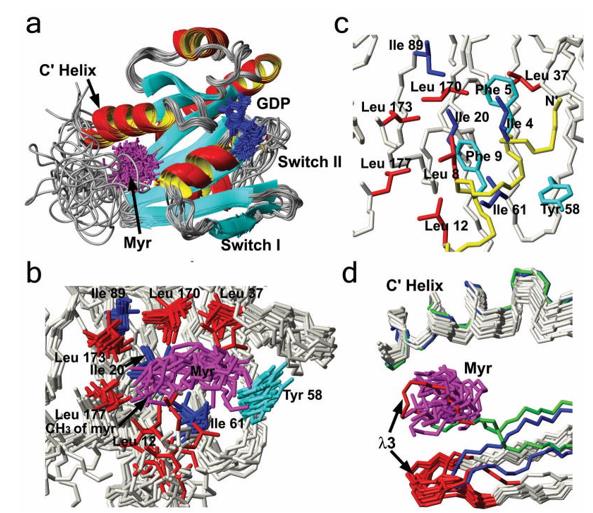 Figure 1. Structure of myristoylated yARF1 and comparisons to related structures. (Liu Y, et al., 2009)
Figure 1. Structure of myristoylated yARF1 and comparisons to related structures. (Liu Y, et al., 2009)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Myirstoylated yeast ARF1 protein, GDP-bound (expressed in E. coli) | Saccharomyces cerevisiae | Solution NMR | / | 2K5U |
| ADP-ribosylation factor (ARF1*GTP), myristoylated (expressed in E. coli) | Saccharomyces cerevisiae | Solution NMR | / | 2KSQ |
Table 1. Structural Research of ADP-Ribosylation Factors.
Structural analysis of transmembrane domains of membrane proteins is important for designing drugs and understanding biological processes. Solution NMR spectroscopy techniques can be used for structural analysis of transmembrane structural domains, thus providing critical information for studying membrane protein functions and developing potential therapeutic strategies.
Solution NMR spectroscopy offers great advantages. In solution, the fast-tumbling motion of molecules eliminates various anisotropic NMR interactions that may broaden the spectral lines. As a result, the resonance signals in liquid NMR spectra are very sharp and have high resolution, which is one of the reasons why solution NMR spectroscopy is the most powerful method for determining the structure of membrane proteins.
Do you need to use solution NMR to analyze your membrane proteins? Creative Biostructure is a reliable NMR solution provider. We have advanced NMR equipment and an experienced technical team. Our aim is to build tailor-made protein analysis solutions to obtain accurate and reliable structural information to fuel our customers' research. If you are interested in learning more about our protein structural analysis services, please contact us for more information.
References
- Liu Y, et al. Structure and membrane interaction of Myristoylated ARF1. Structure, 2009, 17(1): 79–87.
- Liu Y, et al. Dynamic structure of membrane-anchored ARF•GTP. Nature Structural & Molecular Biology, 2010, 17(7): 876–881.