Structural Research of N-acyltransferases
N-acetyltransferases (NATs) are polymorphic drug metabolizing enzymes used to acetylate arylamine carcinogens and drugs. The two NAT enzymes in humans are NAT1 and NAT2, which are particularly important for drug metabolism. NAT2 is also a key marker for diabetes and a cardiovascular risk factor. Therefore, structural studies of NAT to facilitate the development of its inhibitors are critical for a variety of human diseases.
Advances in the structural study of NAT
The crystal structure of Salmonella typhimurium NAT is determined using selenomethionine (SeMet) labeling and multi-wavelength anomalous diffraction (MAD) technique with a resolution of 2.8 Å. The results show a catalytic triplet structure consisting of an α-structural domain, a β-structural domain, and an α/β C-terminal structural domain. The Cys, His, and Asp active sites are located within the α and β domains. The third structural domain folds over the active site and is used to recognize the substrate. The presence of the catalytic triad is essential for activity in all NATs.
Research progress on the action mechanism of NAT
Early kinetic analysis of N-acetylation catalyzed by arylamine showed that the reaction is carried out through the "Ping Pong Bi-Bi" mechanism. This mechanism first causes the enzyme to be acetylated by acetyl donors, resulting in stable acetylase intermediates. The acetyl acceptor is then deacetylated. In the absence of an acetyl acceptor substrate, the acetylase intermediate can also be hydrolyzed.
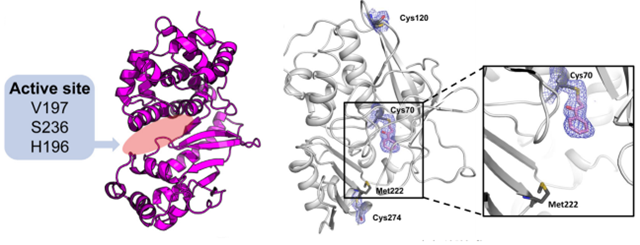 Figure 1. The ribbon diagram of NAT (left) and inhibitor binding to Cys residues in the active site of NAT (right). (Reynolds JA, et al., 2023; Sim
E, et al., 2014)
Figure 1. The ribbon diagram of NAT (left) and inhibitor binding to Cys residues in the active site of NAT (right). (Reynolds JA, et al., 2023; Sim
E, et al., 2014)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Ta0374 in complex with acetyl-coa | Thermoplasma acidophilum | X-ray diffraction | 2.5 Å | 3F0A |
| Acetyltransferase, Tb11.01.2886 | Trypanosoma brucei | X-ray diffraction | 2.35 Å | 3FB3 |
| N-acetyltransferase | Staphylococcus aureus subsp. aureus Mu50 | X-ray diffraction | 1.6001 Å | 4LUA |
| N-acetyltransferase | Staphylococcus aureus subsp. aureus Mu50 | X-ray diffraction | 2 Å | 4M85 |
| Apo AtaTR | Escherichia coli | X-ray diffraction | 2.604 Å | 6AJM |
| AtaTR bound with AcCoA | Escherichia coli | X-ray diffraction | 3.302 Å | 6AJN |
| AtaT Y144F mutant toxin | Escherichia coli | X-ray diffraction | 2.5 Å | 6GTP |
| AtaT Y144F mutant toxin bound to the C-terminus of the antitoxin AtaR | Escherichia coli | X-ray diffraction | 2.49 Å | 6GTQ |
| AtaT Y144F mutant toxin bound to the C-terminus of the antitoxin AtaR and Acetyl-CoA | Escherichia coli | X-ray diffraction | 2.99 Å | 6GTR |
| N-acetyltransferase in the presence of Coenzyme A and dTDP-3-amino-3,6-dideoxy-D-glucose | Helicobacter pullorum | X-ray diffraction | 1.45 Å | 7S3U |
| Acetyltransferase effector VipF | Legionella pneumophila | X-ray diffraction | 2.392 Å | 7WX5 |
| Acetyltransferase VipF and COA/ACO | Legionella pneumophila | X-ray diffraction | 1.781 Å | 7WX7 |
| GCN5-related N-Acetyltransferase | Lactobacillus | X-ray diffraction | 1.95 Å | 8OSP |
| N-acetyltransferase | Staphylococcus aureus | X-ray diffraction | 1.81 Å | 5IX3 |
| SpeG spermidine N-acetyltransferase in complex with spermine | Staphylococcus aureus | X-ray diffraction | 2.65 Å | 8FV0 |
| NAT1 (FDB2) N-malonyltransferase | Fusarium verticillioides | X-ray diffraction | 1.8 Å | 7QI3 |
| Arylamine N-acetyltransferase F42W mutant | Mesorhizobium japonicum MAFF 303099 | X-ray diffraction | 1.84 Å | 4NV8 |
| Arylamine N-acetyltransferase 1 In Complex With CoA | Mesorhizobium japonicum MAFF 303099 | X-ray diffraction | 2.02 Å | 4NV7 |
| Arylamine N-acetyltransferase C | Bacillus anthracis | X-ray diffraction | 2.01 Å | 3LNB |
| N-acetyltransferase 1 | Homo sapiens | X-ray diffraction | 1.78 Å | 2PQT |
| N-acetyltransferase 2 | Homo sapiens | X-ray diffraction | 1.92 Å | 2PFR |
| (BACCR)NAT3 arylamine N-acetyltransferase reveals a unique Cys-His-Glu catalytic triad | Bacillus cereus ATCC 14579 | X-ray diffraction | 2.14 Å | 4DMO |
| N-terminal acetyltransferase NatE (IP6) in complex with a bisubstrate | Saccharomyces cerevisiae | X-ray diffraction | 2.1 Å | 4XNH |
| Arylamine N-acetyltransferase 1 | Mesorhizobium loti | X-ray diffraction | 2 Å | 2BSZ |
Table 1. Structural research of N-acyltransferases.
To study the structure of N-acyltransferases, X-ray crystallography is commonly used. Structural analysis of NAT proteins can help develop drugs for diabetes and cancer.
Creative Biostructure has long been committed to the study of structural biology and membrane proteins. We have extensive experience in determining membrane protein structures.
In addition to the structural determination of membrane proteins, we can accurately analyze biomolecules, including but not limited to nucleic acids, ribosomes, small proteins, protein complexes, protein-ligand complexes, and viruses. If you are interested in our services, please contact us for more details.
References
- Reynolds JA, et al. An engineered N-acyltransferase-LOV2 domain fusion protein enables light-inducible allosteric control of enzymatic activity. J Biol Chem. 2023.299(4):103069.
- Sim E, et al. Arylamine N-acetyltransferases: from drug metabolism and pharmacogenetics to drug discovery. Br J Pharmacol. 2014.171(11):2705-2725.
- Sinclair, J., et al. Structure of arylamine N-acetyltransferase reveals a catalytic triad. Nat Struct Mol Biol 7. 2000:560–564.