Structural Research of Solute Carrier Family 4
The solute carrier (SLC) superfamily currently includes 458 transport proteins from 65 families that carry a wide range of substances across cell membranes. Human SLCs transport nutrients, metabolites, exogenous substances, and drugs, including inorganic ions, amino acids, sugars, neurotransmitters, lipids, and drugs. Most SLCs function as coupled co-transporter proteins that utilize the H+ or Na+ gradient as a driving force for the transport of substrates against the concentration gradient into the cell, and play essential roles in various physiological and pharmacological processes.
General structure analysis of SLC4 family transporter proteins
SLC4 family transporters are N-glycosylated integrated membrane proteins. Research has found that the length and molecular weight of peptides vary greatly among different members and even variants of the SLC4 family. The length of the entire peptide of SLC4 members ranges from approximately 850 to 1250 amino acids. The SLC4 protein typically contains three main structural domains, the intracellular amino-terminal (Nt) domain; Multi transmembrane domain (TMD); Cellular carboxyl-terminal (Ct) domain.
Structural differences of SLC4 proteins in different models
The first SLC4 family member is identified from erythrocytes as AE1. Due to the inability of cryo-electron microscopy to determine the topological structure of TMD, researchers' understanding of the SLC4 protein structure largely stems from a series of biochemical studies on AE1 and NBCe1. Two models are proposed regarding the TMD topology of SLC4 protein. In Model A, 13 of the 14 TMs (except TM12) are predicted to be helical, whereas all 14 TMs in Model B are predicted to be helical. The two models differed between TM9 and TM13. The reentrant loop RL1 between TM9 and TM10 in model A forms a transmembrane helix (TM10) and an intracellular loop (IL5) in model B. The reentrant loop RL1 between TM9 and TM10 in model A forms a transmembrane helix (TM10) in model B.
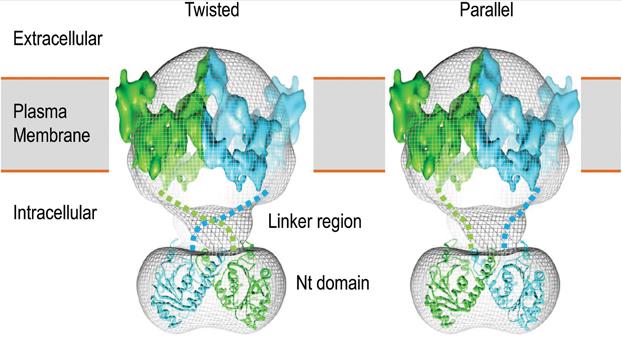 Figure 1. 3D structural model of human AE1 dimer. (Liu Y, et al., 2015)
Figure 1. 3D structural model of human AE1 dimer. (Liu Y, et al., 2015)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| bAE1 captured in multiple states. | Bos taurus | Cryo-EM single particle analysis | 4.4 Å | 8D9N |
| bAE1 captured in multiple states | Bos taurus | Cryo-EM single particle analysis | 6 Å | 8E34 |
| bAE1 captured in multiple states | Bos taurus | Cryo-EM single particle analysis | 6.3 Å | 8EEQ |
| Fab4201 raised against Human Erythrocyte Anion Exchanger 1 | Mus musculus | X-ray diffraction | 2.5 Å | 5A16 |
| The anion exchanger domain of human erythrocyte Band 3 | Homo sapiens | X-ray diffraction | 3.5 Å | 4YZF |
| A low energy structure for the final cytoplasmic loop of band 3 | Homo sapiens | SOLUTION NMR | / | 1BH7 |
| The first and second transmembrane-spanning segments of band 3 | Homo sapiens | SOLUTION NMR | / | 1BTR |
| The cytoplasmic domain of erythrocyte band-3 protein | Mus musculus | X-ray diffraction | 2.6 Å | 1YHN |
| The intermediate structure of hAE2 in basic pH | Homo sapiens | Cryo-EM single particle analysis | 3.25 Å | 8GVA |
| hAE2 with DIDS | Homo sapiens | Cryo-EM single particle analysis | 3.08 Å | 8GV8 |
| hAE2 with chloride ion | Homo sapiens | Cryo-EM single particle analysis | 3.06 Å | 8GV9 |
| hAE2 with bicarbonate | Homo sapiens | Cryo-EM single particle analysis | 2.89 Å | 8GVC |
| hAE2 | Homo sapiens | Cryo-EM single particle analysis | 3.17 Å | 8GVE |
| hAE2 in basic pH | Homo sapiens | Cryo-EM single particle analysis | 3.09 Å | 8GVF |
| AE2 in acidic KNO3 | Homo sapiens | Cryo-EM single particle analysis | 3.32 Å | 8GVH |
| SLC4A4 sodium-coupled acid-base transporter NBCe1 | Homo sapiens | Cryo-EM single particle analysis | 3.9 Å | 6CAA |
| SLC4 transporter Bor1p in an inward-facing conformation | Saccharomyces mikatae | Cryo-EM single particle analysis | 5.9 Å | 5SV9 |
| Sodium-driven chloride/bicarbonate exchanger NDCBE (SLC4A8) | Oryctolagus cuniculus | Cryo-EM single particle analysis | 3.4 Å | 7RTM |
| The regulatory domain of the sodium-driven chloride bicarbonate exchanger. | Homo sapiens | X-ray diffraction | 2.801 Å | 5JHO |
| BTR1 in the inward-facing state at pH 5.5 | Homo sapiens | Cryo-EM single particle analysis | 2.94 Å | 7X1G |
| BTR1 in the inward-facing state with R125H mutation | Homo sapiens | Cryo-EM single particle analysis | 2.96 Å | 7X1H |
| BTR1 in the outward-facing state | Homo sapiens | Cryo-EM single particle analysis | 2.94 Å | 7X1I |
| BTR1 in the outward-facing state in the presence of NH4Cl | Homo sapiens | Cryo-EM single particle analysis | 2.84 Å | 7X1J |
Table 1. Structural research of the solute carrier family 4.
Research has shown that SLC4 proteins are considered tumor therapeutic targets in clinical trials. Through the structural analysis of SLC4, its function and action mechanism will be further explored to discover the potential clinical prospects of SLC4 protein.
Creative Biostructure, a leader in structural research, offers protein structure analysis services to help researchers study the structure and function of SLC4 and other complex proteins. Our services include X-ray crystallography, cryo-electron microscopy (cryo-EM), and nuclear magnetic resonance (NMR) spectroscopy. If you are interested in exploring the structure of proteins and would like to learn more about our services, please feel free to contact us. Our team is always ready to discuss your research requirements and provide you with the best solution for your project.
References
- Liu Y, et al. Structure and Function of SLC4 Family [Formula: see text] Transporters. Front Physiol. 2015. 6: 355.
- Romero MF, et al. The SLC4 family of bicarbonate (HCO3-) transporters. Mol Aspects Med. 2013. 34(2-3): 159-182.
- Dong J, et al. [Research progress on structure, function and disease correlation of solute carrier family 4]. Sheng Li Xue Bao. 2023.75(1): 137-150.