Structural Research of Potyviridae
The Potyviridae are a family of positive-stranded RNA viruses belonging to the small nucleovirus supergroup, which cause significant economic losses to a wide range of crops and have essential agricultural and ecological impacts. Therefore, Potyviridae have received more research than other plant viruses. In past research, to better understand potyvirus and its infection process, researchers have structurally characterized potyvirus using high-resolution cryo-electron microscopy (cryo-EM) as well as cutting-edge small-angle X-ray scattering (SAXS) and atomic force microscopy (AFM) techniques.
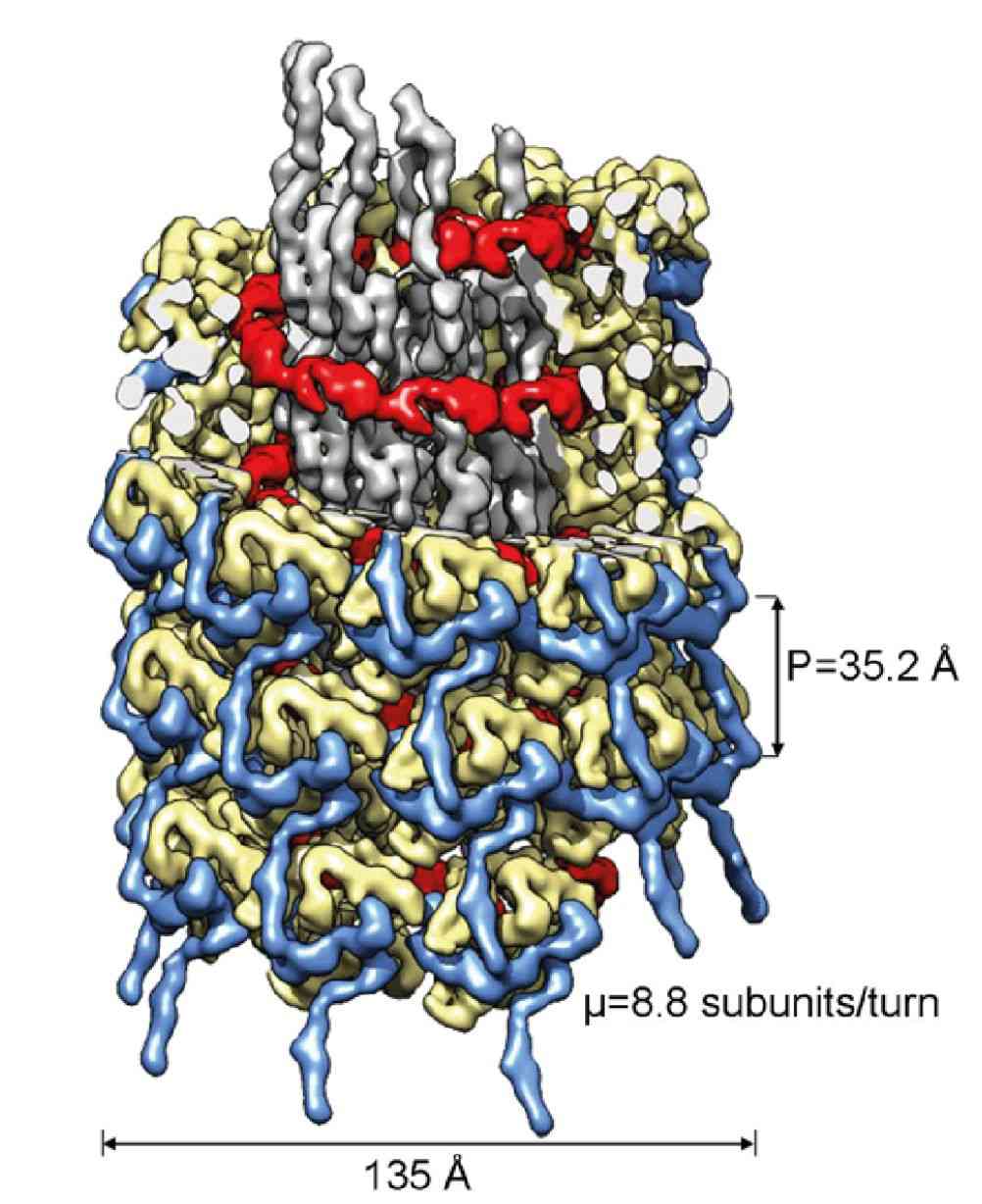 Figure 1. Cryo-electron microscopy of the turnip mosaic virus (TuMV), a member of Potyviridae. (Inoue-Nagata AK, et al, 2022)
Figure 1. Cryo-electron microscopy of the turnip mosaic virus (TuMV), a member of Potyviridae. (Inoue-Nagata AK, et al, 2022)
Progress in the Structural Resolution of Potyviridae
In contrast to rigid rod-shaped plant viruses, X-ray crystallography is not suitable for resolving the 3D structure of filamentous plant viruses (e.g. potyviruses). A wealth of information for structural research comes from X-ray fiber diffraction and electron microscopy (EM). At the beginning of the 21st century, transmission electron microscopy (TEM) was used to determine the shape and size of various virus particles. This was followed by the use of negative staining to construct virus bodies. Since many helical plant viruses could not be resolved by classical structural methods, SAXS and AFM matured and became valuable tools for researching various potyviruses.
Structural Features of Potyviridae Family Members
Sweet potato feathery mottle virus (SPFMV) and sweet potato mild mottle virus (SPMMV) are typical members of the Potyviridae family. Researchers generated virus-like particles (VLP) by transient expression of the capsid protein (CP) of SPFMV and SPMMV and analyzed the purified VLP by cryo-electron microscopy, which yielded structures with resolutions of 2.6 and 3.0 Å, respectively, showing a left-handed helix-like arrangement of 8.8 CP subunits per circle. The core domain consists of nine α helices and two β strands. The RNA-binding pocket is located at the gap formed by the core domain and the C-terminal tail. The C-terminal ends of the filaments form a helical arrangement, resulting in a dense inner tube formed by the most C-terminal end of the CP.
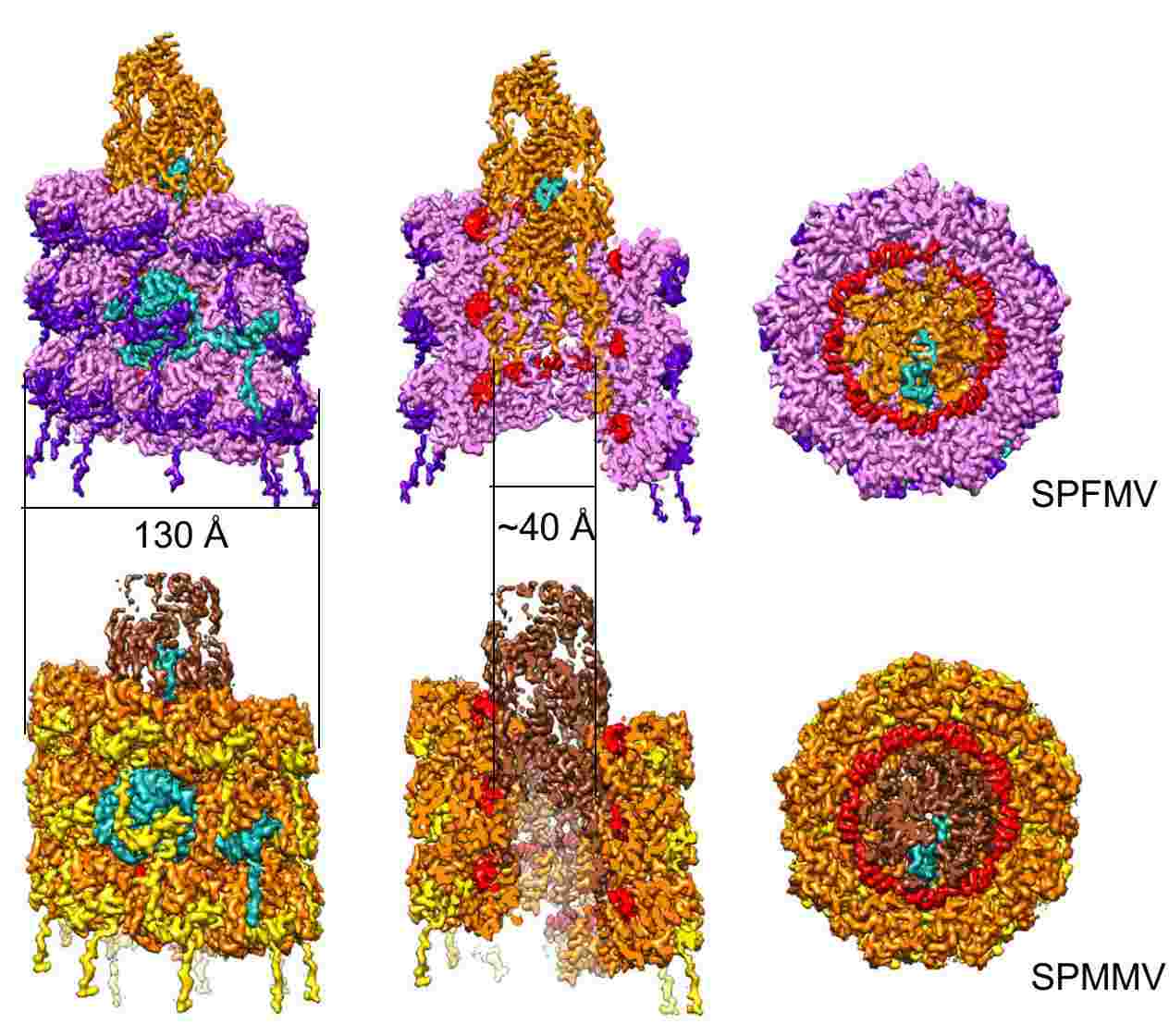 Figure 2. Cryo-electron microscopy of SPFMV and SPMMV VLPs. (Chase O, et al, 2023)
Figure 2. Cryo-electron microscopy of SPFMV and SPMMV VLPs. (Chase O, et al, 2023)
Helical plant viruses can be used in bio-nanotechnology to produce heterologous proteins in plants for vaccine production or drug delivery. Some helical plant viruses are considered promising tools for the development of nanomaterials in modern medicine, microelectronics, and a variety of fields. Creative Biostructure is a biomolecular structure analysis company specializing in virus-like particles (VLPs). Our wide range of VLP products satisfies the requirements of researchers, vaccine developers, and diagnostic professionals and is an ideal platform for research into virus entry, immunology, and vaccine development.
| Cat No. | Product Name | Virus Name | Source | Composition |
| CBS-V720 | Chimeric PhMV & JEV VLP (CP; JEV E protein(epitopes) Proteins) | Physalis mottle virus; Japanese encephalitis virus | E. coli recombinant | CP (Coat Protein); JEV E protein(epitopes) |
| Explore All Potyviridae Virus-like Particle Products | ||||
Understanding the structure of viruses at atomic resolution is essential for the discovery and design of vaccines and antiviral drugs, as well as drug delivery vectors. Creative Biostructure, a leader in the structural resolution of biomacromolecules, offers cutting-edge cryo-electron microscopy (cryo-EM) to assist clients in determining the high-resolution three-dimensional structure of viral particles and virus-like particles. If you are interested in our services and VLP products, please feel free to contact us for more information.
References
- Inoue-Nagata AK, et al. ICTV Virus Taxonomy Profile: Potyviridae 2022. J Gen Virol. 2022. 103(5): 10.1099/jgv.0.001738.
- Chase O, et al. CryoEM and stability analysis of virus-like particles of potyvirus and ipomovirus infecting a common host. Commun Biol. 2023. 6(1): 433.
- Shtykova EV, et al. Structural Insights into Plant Viruses Revealed by Small-Angle X-ray Scattering and Atomic Force Microscopy. Viruses. 2024. 16(3): 427.