Structural Research of Variant Surface Glycoproteins (VSG)
Variant surface glycoproteins (VSGs) are proteins with a size of about 60 kDa, tightly wrapped on the cell surface of protozoan parasites belonging to the genus Trypanosoma. Such proteins are highly immunogenic, and the immune response to a specific VSG shell will rapidly kill trypanosomes expressing this variant. They play key functions and protective roles on the surface of these microorganisms. The structural findings of VSGs revealed key details of their interactions with the host immune system. By analyzing the structure and function of these proteins, it is of great significance for the study of related diseases and vaccine development.
VSG consists of an N-terminal domain of about 300-350 amino acids with low sequence homology and a more conserved C-terminal domain of about 100 amino acids. The N-terminal domains are classified into AC classes according to their cysteine patterns. The C-term domain is divided into classes I-III according to sequence homology. For dimerization, the VSG N-terminal domain forms a bundle consisting of four α A bundles of helices surrounded by smaller structural features (five smaller helices and three β Fold). Scientists used small-angle X-ray scattering to elucidate the complete VSG structure. The resulting model shows that the linker region confers great flexibility between domains, suggesting that VSG can adopt two major conformations to cope with the changes in barrier and protein density while always maintaining a protective barrier.
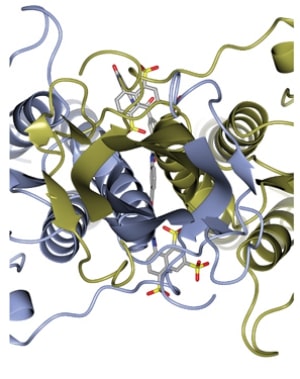 Figure 1. Ribbon diagram of VSGsur (one monomer blue and the other gold) looking down the two-fold axis of symmrtry for the dimer.
(Zeelen J, et al., 2021)
Figure 1. Ribbon diagram of VSGsur (one monomer blue and the other gold) looking down the two-fold axis of symmrtry for the dimer.
(Zeelen J, et al., 2021)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| N-terminal Domain of VSG2 (MITat 1.2): Trypanosoma brucei | Trypanosoma brucei brucei | X-ray diffraction | 2.90 Å | 1VSG |
| N-terminal of VSG ILTat 1.24: Trypanosoma brucei | Saccharomyces cerevisiae | X-ray diffraction | 2.70 Å | 2VSG |
| N-terminal domain of VSG M1.1: Trypanosoma brucei | Trypanosoma brucei | X-ray diffraction | 1.65 Å | 5LY9 |
| N-terminal Domain of VSG3: Trypanosoma brucei | Mus musculus | X-ray diffraction | 1.41 Å | 6ELC |
| Variant Surface Glycoproteins VSGsur & VSG13: Trypanosoma brucei | Trypanosoma brucei rhodesiense | X-ray diffraction | 1.21 Å | 6Z7A |
Table 1. Structural research of variant surface glycoproteins.
The structural biology methods and techniques used in the structural study of VSGs, such as X-ray crystallography, are to obtain the crystals of VSGs and use X-ray diffraction to determine their high-resolution three-dimensional structure. Nuclear magnetic resonance (NMR) technology can obtain the dissociation constant, kinetic parameters, and conformational information of VSGs labeled proteins through NMR experiments. Using single particle cryo-electron microscopy technology, the structural information of VSGs under physiological conditions can be analyzed.
Creative Biostructure is a biological company that focuses on protein structural analysis. We provide a wide range of protein structure analysis services and resources for scientists and researchers. It includes VSGs crystallization screening and optimization and helps customers obtain high-quality VSGs crystallization samples through the most advanced crystallization screening technology. High-resolution X-ray crystallography was used to determine the three-dimensional structure of VSGs. And use existing structural information to predict the structure and function of VSGs. If you want to know more about the types of services we provide, please feel free to contact us.
References
- Zeelen J, et al. Structure of trypanosome coat protein VSGsur and function in suramin resistance. Nat Microbiol. 2021; 6 (3): 392-400.
- Freymann D, et al. 2.9 Å resolution structure of the N-terminal domain of a variant surface glycoprotein from Trypanosoma brucei. Journal of Molecular Biology. 1990. 216 (1): 141–60.