Structural Research of Shape, Elongation, Division, and Sporulation (SEDS) Proteins
SEDS (shape, elongation, division, and sporulation) proteins are broadly conserved peptidoglycan (PG) glycosyltransferases that form a complex with class B penicillin-binding proteins (bPBPs) to synthesize cell wall PGs during bacterial growth and division. Since SEDS proteins play essential roles in bacterial morphogenesis, the elucidation of their structures and action mechanisms is one of the promising avenues for the development of novel antibiotics.
SEDS protein structure analysis method
SEDS glycosyltransferase and class B penicillin-binding protein (PBP) form the core of the multiprotein complex required for PG assembly. In E. coli, PBP2 and RodA from the SEDS family mediate PG formation. Using single-particle cryo-electron microscopy, the researchers determined the structure of the RodA-PBP2 complex, which was purified to homogeneity by metal affinity chromatography and SEC in detergent, expressed as a functional fusion and reconstructed in lipid-filled nanodiscs.
Structural analysis of SEDS proteins
The structure indicates that RodA is an intact membrane protein consisting of 10 transmembrane (TM) helices. PBP2 has a single TM helix and an extracellular structural domain with classical class B PBP folds. In TM mapping, RodA is shown to have an intracellular N-terminus and a C-terminus.TM helices 1-6 and TM helices 8-10 form a tight helical bundle, and TM helix 7 extends away from it and is stabilized by three periplasmic near-membrane helices (PH1, PH2, and PH3). The PBP2 single TM helices are packed on RodA TM8 and TM9.
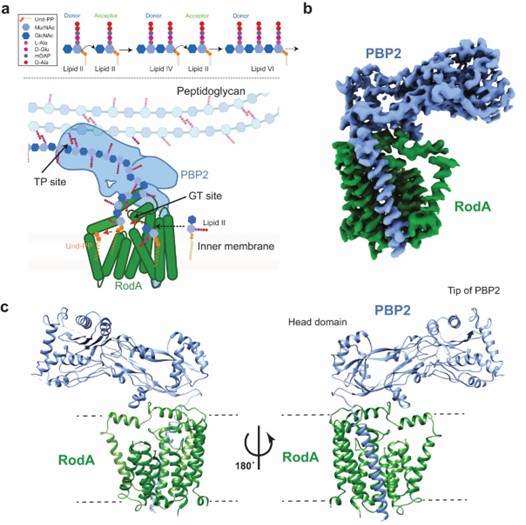 Figure 1. Mechanism and overall structure of RodA-PBP2 complex. (Tsukazaki T, 2019)
Figure 1. Mechanism and overall structure of RodA-PBP2 complex. (Tsukazaki T, 2019)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Rod shape determining protein RodA (Q5SIX3_THET8) | Thermus thermophilus HB8 | X-ray diffraction | 2.908 Å | 6BAR |
| Rod shape determining protein RodA D255A mutant (Q5SIX3_THET8) | Thermus thermophilus HB8 | X-ray diffraction | 3.194 Å | 6BAS |
| Polymerization and crosslinking by a peptidoglycan synthase complex | Thermus thermophilus HB8 | X-ray diffraction | 3.5 Å | 6PL5 |
| Penicillin-binding protein 3 | Bacillus subtilis subsp. subtilis str. 168 | X-ray diffraction | 2.65 Å | 7BN9 |
| Rho1 | Schizosaccharomyces pombe 972h- | X-ray diffraction | 2.78 Å | 8ETD |
| PBP1b with a FtsN peptide activating transglycosylase activity | Escherichia coli K-12 | X-ray diffraction | 2.4 Å | 6YN0 |
| Rho1 F25N | Aspergillus fumigatus Af293 | X-ray diffraction | 1.42 Å | 5ZVP |
| Sec4p, a Rab family GTPase | Candida albicans SC5314 | X-ray diffraction | 1.88 Å | 6O62 |
| Core-mannan synthase A (CmsA/Ktr4), Mn/GDP-form | Aspergillus fumigatus A1163 | X-ray diffraction | 1.9 Å | 7BOP |
| Rab GTPase Sec4p mutant - S29V | Saccharomyces cerevisiae S288C | X-ray diffraction | 1.9 Å | 4Z8Y |
| CDC10 - CDC3 heterocomplex | Saccharomyces cerevisiae S288C | X-ray diffraction | 2.22 Å | 8FWP |
| CDC3(G) - CDC10(Delta 1-10) heterocomplex | Saccharomyces cerevisiae | X-ray diffraction | 2.66 Å | 8SGD |
| CTD binding of the Set2 SRI domain | Saccharomyces cerevisiae | SOLUTION NMR | / | 2C5Z |
| SEC4-GDP | Saccharomyces cerevisiae | X-ray diffraction | 1.8 Å | 1G16 |
| Sec4 in complex with Rab-GDI | Saccharomyces cerevisiae | X-ray diffraction | 2.9 Å | 3CPH |
| N-terminal domain of GpsB in complex with PBP4 fragment | Staphylococcus aureus subsp. aureus COL | X-ray diffraction | 2.4 Å | 8E2C |
| N-terminal domain of GpsB | Staphylococcus aureus subsp. aureus COL | X-ray diffraction | 1.95 Å | 8E2B |
| CetZ - GDP bound monomer | Haloferax volcanii | X-ray diffraction | 1.9 Å | 4B46 |
| CetZ strain DSM 6194 | Methanothrix thermoacetophila | X-ray diffraction | 2 Å | 3ZID |
| Set2 bound to a nucleosome containing oncohistone mutations | Saccharomyces cerevisiae | Cryo-EM single particle analysis | 3.3 Å | 7EA5 |
| Glc7 phosphatase | Saccharomyces cerevisiae S288C | X-ray diffraction | 1.65 Å | 7QWJ |
| Glc7 phosphatase in complex with the regulatory region of Ref2 | Saccharomyces cerevisiae | X-ray diffraction | 1.85 Å | 8A8F |
Table 1. Structural research of shape, elongation, division, and sporulation (SEDS) proteins.
Creative Biostructure is a leading provider of structural biology services that help researchers deepen their understanding of complex protein structures. We offer a range of technologies to help determine the structure of shape, elongation, division, and sporulation (SEDS) proteins, including X-ray crystallography, cryo-electron microscopy (cryo-EM), and nuclear magnetic resonance (NMR). Through our services, researchers can better utilize SEDS proteins to develop novel antibiotics.
If you are interested in exploring the structure of SEDS proteins or other membrane proteins, please feel free to contact us. We offer a comprehensive one-stop structural biology service ranging from protein expression and purification to structure determination.
References
- Nygaard R, et al. Structural basis of peptidoglycan synthesis by E. coli RodA-PBP2 complex. Nat Commun. 2023. 14(1): 5151.
- Li Y, et al. Identification of the potential active site of the septal peptidoglycan polymerase FtsW. PLoS Genet. 2022. 18(1): e1009993.
- Sjodt M, et al. Structural coordination of polymerization and crosslinking by a SEDS-bPBP peptidoglycan synthase complex. Nat Microbiol. 2020. 5(6): 813-820.