Structural Research of Methyltransferases
Methyltransferase is an enzyme that transfers a methyl group from S-adenosine methionine (SAM) to its substrate to methylate it. These enzymatic reactions are present in many pathways and have been implicated in genetic diseases, cancers, and metabolic diseases. It can be divided into several subclasses according to its structural characteristics. The most common is Class I, which is used to combine SAM's Rossman folds. Class II contains SET domains, and Class III methyltransferases are membrane-related.
Class III Methyltransferase Structural Analysis
Class III methyltransferase is a dimer with an active site located between two domains and a similar composition of lobes, both with a central β fragment and an alpha-helix on either side. Their topologies are different, the C-terminal domain has a β-turn between chain 4 and chain 5, thus forming an anti-parallel fragment, and the protein dimerizes on the bridge between the two domains. Using a tightly folded conformation, the activated methyl group is exposed to the outside.
Research Progress of Membrane-related Methyltransferase
Studies have shown that methyltransferase activity is mainly concentrated in the endoplasmic reticulum and mitochondria-associated membranes. The membrane-associated trimer complex VapA-VipC-VapB controls the signal transduction pathway of fungal differentiation. VipC-VapB methyltransferase is attached to the membrane via the Fyve-like zinc finger protein VapA, thus allowing the nuclear VelB-VeA-LaeA complex to activate transcription to promote sexual development.
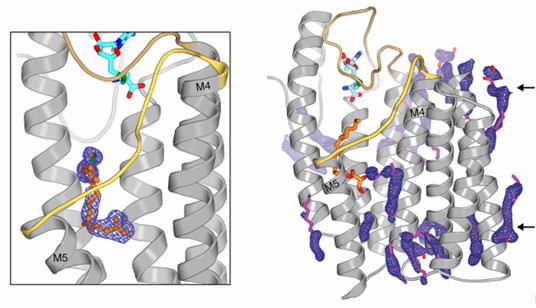 Figure 1. Electron density for the active site and monoolein lipids surround the transmembrane region of ICMT. (Diver, M.
et al., 2018)
Figure 1. Electron density for the active site and monoolein lipids surround the transmembrane region of ICMT. (Diver, M.
et al., 2018)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| PKMT1 | Rickettsia prowazekii str. Madrid E | X-ray diffraction | 2.60 Å | 5DO0 |
| PKMT1 in complex with AdoHcy | Rickettsia prowazekii str. Madrid E | X-ray diffraction | 1.90 Å | 5DNK |
| Putative O-methyltransferase | Halalkalibacterium halodurans C-125 | X-ray diffraction | 1.90 Å | 2GPY |
| Isoflavone O-methyltransferase homolog in complex with biochanin A and SAH | Medicago truncatula | X-ray diffraction | 1.95 Å | 2QYO |
| PFOMT, Phenylpropanoid, and Flavonoid O-methyltransferase | Mesembryanthemum crystallinum | X-ray diffraction | 1.371 Å | 3C3Y |
| Cationdependent O-Methyltransferase. | Synechocystis sp. PCC 6803 | X-ray diffraction | 2.0 Å | 3CBG |
| O-methyltransferase (NcsB1) in complex with S-adenosyl-L-homocysteine (SAH) | Streptomyces carzinostaticus subsp. neocarzinostaticus | X-ray diffraction | 2.08 Å | 3I53 |
| O-methyltransferase family protein | Burkholderia thailandensis E264 | X-ray diffraction | 1.90 Å | 3MCZ |
| PKMT2 | Rickettsia typhi str. Wilmington | X-ray diffraction | 3.133 Å | 5DOO |
| PKMT2 in complex with AdoHcy | Rickettsia typhi str. Wilmington | X-ray diffraction | 3.20 Å | 5DPL |
| Glycine sarcosine N-methyltransferase in complex with betaine | Methanohalophilus portucalensis FDF-1 | X-ray diffraction | 1.93 Å | 5HIJ |
| Chalcone o-methyltransferase | Medicago sativa | X-ray diffraction | 1.82 Å | 1FP1 |
Table 1. Structural research of methyltransferases.
Creative Biostructure uses X-ray crystallography to investigate membrane-related methyltransferase. The obtained cryo-electron microscopy (cryo-EM) structure is conducive to the study of its function and role in disease and could help the development of targeted drugs.
We have long been committed to the study of structural biology and membrane proteins. Our experts have extensive experience in the determination of membrane protein structures.
In addition to the structural determination of membrane proteins, we can accurately analyze other biomolecules, including but not limited to ribosomes, nucleic acids, small proteins, protein complexes, protein-ligand complexes, and viruses. If you are interested in our services, please contact us for more details.
References
- Diver, M. et al. Atomic structure of the eukaryotic intramembrane RAS methyltransferase ICMT. Nature, 2018,553: 526–529.
- Sarikaya-Bayram O et al. Membrane-bound methyltransferase complex VapA-VipC-VapB guides epigenetic control of fungal development. Dev Cell. 2014, 27;29(4):406-420.
- Shields DJ, et al. Membrane topography of human phosphatidylethanolamine N-methyltransferase. J Biol Chem. 2003, 31;278(5):2956-2962.