Screening/Selection Strategies for Protein Engineering
Creative Biostructure has developed the comprehensive protein evolution and engineering platform, aiming at offering world-leading high-throughput screening/selection services for protein evolution and protein engineering. Genetic screening requires the inspection of individual phenotypes, which can be used to discover genes associated with specific phenotypes. On the contrary, selection can bypass the requirement to individually inspect each library member and instead link an activity of interest to physical separation of the encoding DNA or to survival of the organism producing active and desired library members.
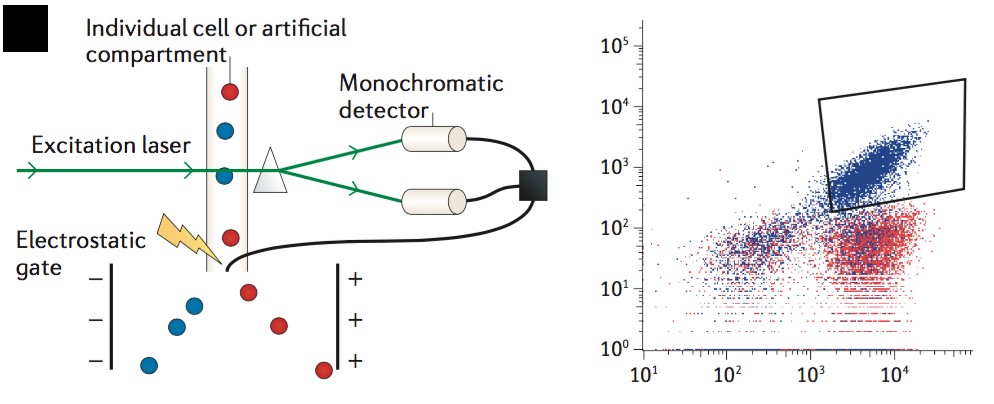 Figure 1. Schematics of Fluorescence-activated cell sorting (FACS) technology. (Nature reviews. Genetics, 2015)
Figure 1. Schematics of Fluorescence-activated cell sorting (FACS) technology. (Nature reviews. Genetics, 2015)
Creative Biostructure can generate abundant protein mutants through outstanding gene diversification libraries, then screening or selection strategies will be an efficient way to obtain desired functional proteins. However, generating a large collection of protein mutants is often time-consuming and labor-intensive. Creative Biostructure’s science team can assist you to speed up your project by providing high-through screening or selection services. We can also provide protein purification and functional assay services.
Creative Biostructure can perform various different strategies for high-throughput screening or selection services, including but not limited to:
Spatial separation is also called colonies screening, which has to take the limitations of screened library size into consideration. The key advantage of this approach is its broad compatibility compared with many other different screening techniques. Additionally, almost any enzymatic activity can be screened in the spatially separated library format.
Using flow cytometry, a bulk population can be interrogated at the level of individual cells using the cell wall or membrane to maintain genotype–phenotype association. Creative Biostructure is able to perform fluorescent activating cell sorting (FACS) based methods to identify and isolate cells containing desired gene variants. Most notably, Creative Biostructure can also use phage display, yeast display, cell-free display, ribosome display and, other cell surface display to screen for complex enzymatic activities.
Compared to cell-constrained fluorescent reporters technology, in vitro compartmentalization (IVC) provides an alternative format to enable high-throughput screening. Seasoned scientists from Creative Biostructure adopt the aqueous droplets in water–oil emulsions to compartmentalize individual genes and gene products along with a surrogate fluorogenic substrate.
In a typical target-binding selection, protein library members with desired binding activity and their encoding DNA sequences are captured using an immobilized target, whereas non-binding library members are washed away.
Creative Biostructure can obtain active library members that enable organisms containing their corresponding genes to survive and replicate. In this technique, antibiotic resistance or auxotroph complementation are the most straightforward activity to evolve using the selective replication of E.coli or S. cerevisiae.
Creative Biostructure can perform in vitro selection for enzymes (such as DNA and RNA polymerases) that directly act on DNA or RNA substrates, including compartmentalized self-replication (CSR) and compartmentalized partnered replication (CPR).
Continuous evolution represents all steps of the protein evolution cycle are performed continuously without manual intervention. Creative Biostructure can utilize phage-assisted continuous evolution (PACE) technique to study the evolution of bacterial genomes and antibiotic resistance.
Creative Biostructure also provides Membrane Protein Platform and Phage Display Platform for your specific projects. Please feel free to contact us for a detailed quote.
References:
M. S. Packer and D. R. Liu (2015). Methods for the directed evolution of proteins. Nature reviews. Genetics, 16: 379-394.