Membrane Protein Dynamics
Membrane proteins play crucial roles in various cellular processes, acting as gateways, receptors, and signal transducers. Understanding the dynamics of membrane proteins – how they move and change conformation within the lipid bilayer – is vital for elucidating their functional mechanisms. Membrane protein dynamics encompass various time scales and involve complex interactions with lipids, other proteins, and small molecular ligands.
What is Membrane Protein Dynamics?
Membrane proteins are embedded within the lipid bilayer of cells, where they perform a plethora of functions essential for cellular homeostasis and communication. These proteins can be categorized into several types, including ion channels, transporters, receptors, and enzymes, each with unique dynamic behaviors tailored to their specific functions.
- Membrane Protein Mobility: Membrane proteins exhibit various types of motion such as lateral diffusion within the plane of the membrane, rotational diffusion around their axis, and conformational changes. Lateral diffusion allows proteins to move within the membrane, influencing processes such as signal transduction and membrane trafficking. Rotational diffusion affects the orientation of protein domains important for function interaction.
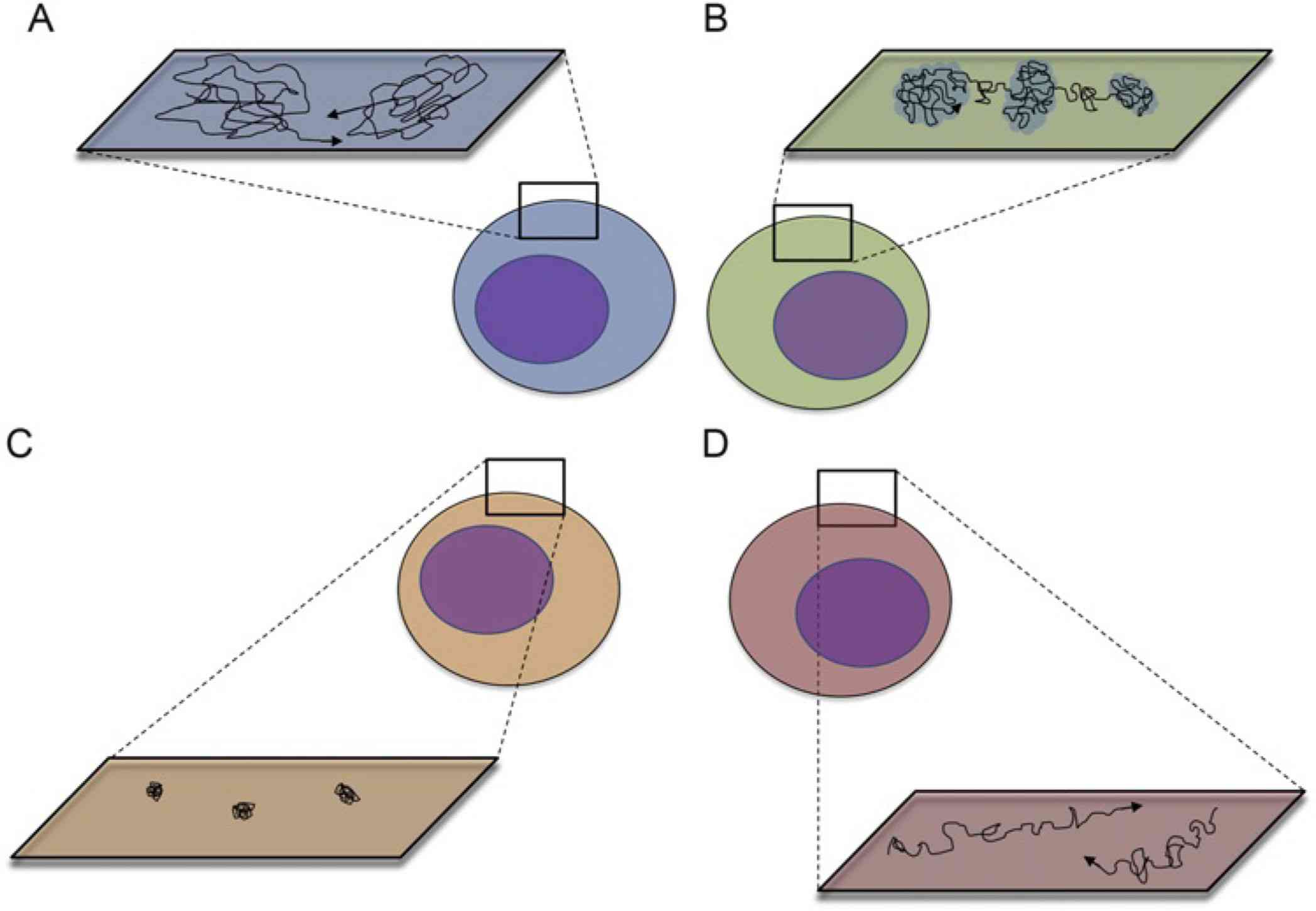 Figure 1. Major modes of membrane protein mobility (A-D): Random Diffusion, Confinement, Restriction, and Directed Motion. (Alenghat F J, et al., 2013)
Figure 1. Major modes of membrane protein mobility (A-D): Random Diffusion, Confinement, Restriction, and Directed Motion. (Alenghat F J, et al., 2013)
- Conformational Changes: Many membrane proteins undergo conformational changes to perform their functions. For example, ion channels open and close in response to voltage changes or ligand binding, transporters switch between different states to move substrates across the membrane, and receptors undergo shape changes upon ligand binding to activate intracellular signaling pathways.
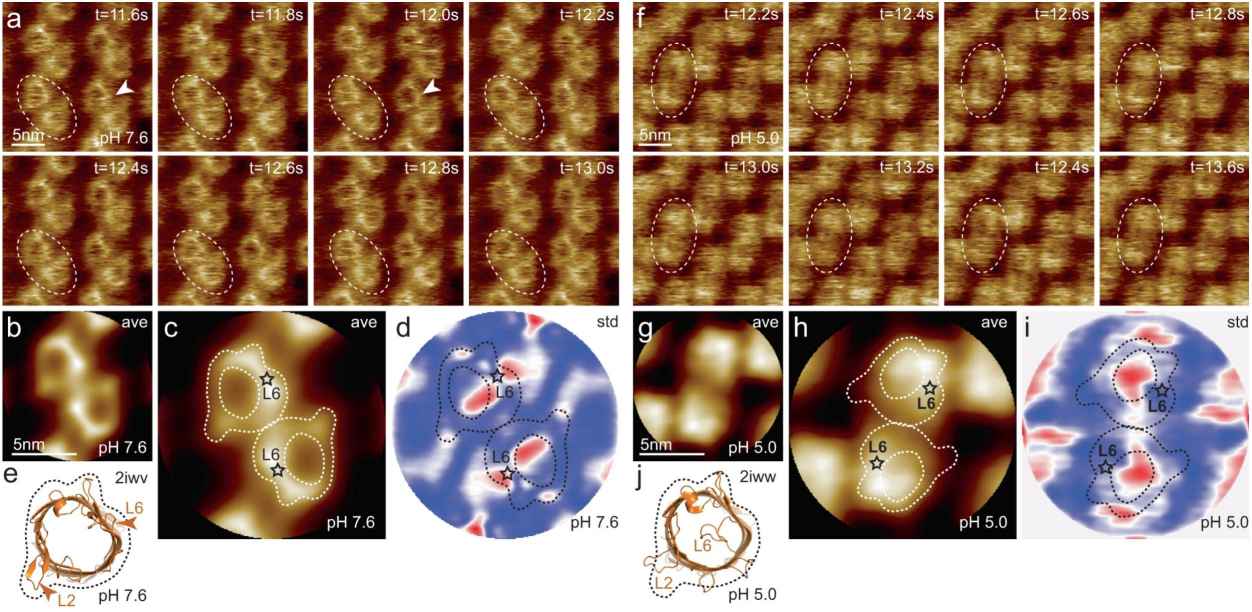 Figure 2. HS-AFM imaging of OmpG in lipid bilayers at pH 7.6 and pH 5.0. (Sanganna Gari R R, et al., 2021)
Figure 2. HS-AFM imaging of OmpG in lipid bilayers at pH 7.6 and pH 5.0. (Sanganna Gari R R, et al., 2021)
Mechanisms Regulating Dynamics
Two primary factors regulate membrane protein dynamics:
- Protein-Protein and Protein-Lipid Interactions: The interactions between membrane proteins and lipids, as well as with other proteins, significantly impact their mobility and function. Specific lipid environments can stabilize certain protein conformations, affecting their activity. Similarly, membrane protein-protein interactions can form large complexes that may restrict or facilitate movement.
- Intracellular and Extracellular Conditions: Factors such as pH, ion concentration, and the presence of specific ligands or inhibitors can modulate the dynamic behavior of membrane proteins. For instance, the binding of a ligand to a receptor can induce conformational changes, altering its activity and mobility within the membrane.
Research Frontiers in Membrane Protein Dynamics
Conformational Plasticity and Allostery
Understanding the conformational flexibility of membrane proteins and the allosteric mechanisms through which distant sites within a protein communicate is a major frontier. Improved insights into allosteric regulation could have profound implications for drug discovery, as many pharmaceutical agents target allosteric sites to modulate protein function with higher specificity and fewer side effects.
Lipid-Protein Interactions
The role of the lipid environment in shaping membrane protein dynamics is another significant area of research. Lipid molecules do not merely serve as a passive matrix but actively participate in modulating the structure, dynamics, and function of membrane proteins. Elucidating these interactions could lead to better understanding of membrane protein function in different physiological and pathological states.
Single-Molecule Dynamics
Studying proteins at the single-molecule level provides a detailed view of their dynamic behaviors that average measurements might obscure. Insights from single-molecule studies can reveal transient states, rare events, and heterogeneity in population behaviors, offering a more comprehensive understanding of membrane protein functions.
Techniques for Studying Membrane Protein Dynamics
Molecular Dynamics Simulations
Molecular dynamics (MD) simulations are a powerful computational technique used to study the motion of atoms and molecules over time, providing detailed insights into the structural dynamics of membrane proteins.
- Principles of MD Simulations: In MD simulations, atoms and molecules are treated as particles governed by Newton's equations of motion. The interactions between particles are described using force fields, which include terms for bond stretching, angle bending, and non-bonded interactions such as van der Waals forces and electrostatic interactions.
- Applications: MD simulations enable the exploration of protein conformations, interactions with ligands and lipids, and the mechanisms of conformational changes. They can simulate processes such as ion channel gating, substrate transport, and receptor activation, providing valuable insights that complement experimental observations.
- Challenges: One of the main challenges of MD simulations is the computational cost, especially for long time-scale events such as large conformational changes. Advances in algorithms and computing power, including the use of specialized hardware like GPUs, are continually improving the feasibility and accuracy of these simulations. At Creative Biostructure, we harness this progress to offer you expert solutions in molecular dynamics.
Fluorescence Spectroscopy
Fluorescence spectroscopy encompasses a range of techniques that use the fluorescent properties of molecules to study the dynamics of membrane proteins.
- Fluorescence Resonance Energy Transfer (FRET): FRET measures the energy transfer between two fluorescently labeled molecules and is sensitive to changes in distance within the range of 1-10 nanometers. This technique is particularly useful for monitoring conformational changes and interactions between protein subunits or between proteins and ligands.
- Fluorescence Recovery After Photobleaching (FRAP): FRAP involves photobleaching a region of fluorescently labeled proteins and measuring the recovery of fluorescence due to the diffusion of unbleached molecules into the bleached area. FRAP provides insights into the lateral mobility of membrane proteins and their interactions with other cellular components.
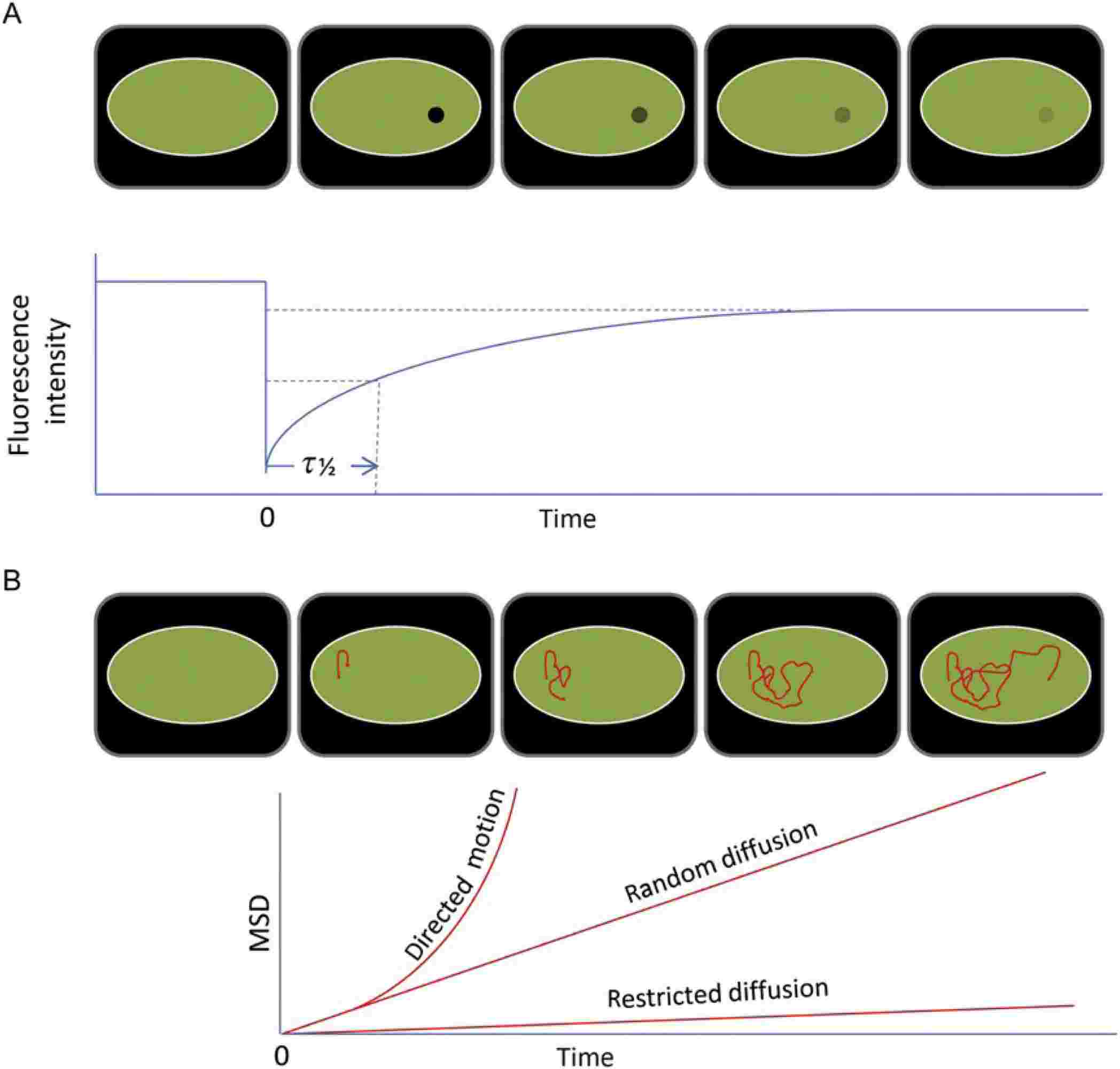 Figure 3. FRAP (A) and SPT (B) measurements are the basis for membrane protein dynamics studies. (Alenghat F J, et al., 2013)
Figure 3. FRAP (A) and SPT (B) measurements are the basis for membrane protein dynamics studies. (Alenghat F J, et al., 2013)
Single-Molecule Techniques
Single-molecule techniques offer unparalleled resolution in studying the dynamics of individual membrane proteins, revealing detailed mechanisms that are often masked in ensemble measurements.
- Single-Molecule Fluorescence Microscopy: This technique allows the observation of single molecules in real-time, providing information on their spatial and temporal behavior. Techniques such as Total Internal Reflection Fluorescence (TIRF) microscopy enhance the signal-to-noise ratio for studying membrane proteins at the cell surface.
- Atomic Force Microscopy (AFM): AFM can measure the topography and mechanical properties of membrane proteins and their complexes. By applying a force and measuring the response, AFM provides insights into the structural integrity and conformational changes of membrane proteins.
- Single-Particle Tracking (SPT): SPT involves tracking the movement of individual particles labeled with fluorescent tags. This technique can reveal detailed information about the diffusion and interactions of membrane proteins within the lipid bilayer, highlighting heterogeneity and dynamic behavior at the single-molecule level.
Whether you require custom membrane protein production or techniques like MD simulations, FRET, FRAP, TIRF microscopy, or AFM, Creative Biostructure has the tools and expertise to support your membrane protein dynamics research. Gain deeper insights into membrane protein functions, interactions, and structural changes. Partner with us to advance your membrane protein studies. Contact us today to learn more.
References
- Alenghat F J, Golan D E. Membrane protein dynamics and functional implications in mammalian cells. Current Topics in Membranes. Academic Press. 2013. 72: 89-120.
- Sanganna Gari R R, Montalvo-Acosta J J, Heath G R, et al. Correlation of membrane protein conformational and functional dynamics. Nature Communications. 2021. 12(1): 4363.