Structural Research of Pro-apoptotic BCL-2 Family
Cellular apoptosis is very important for living organisms. As a key component of the apoptotic pathway, the abnormal function of the Bcl-2 family of proteins can result in the development of a variety of human diseases, including cancer and neurodegenerative diseases. Bcl-2 is mainly located on the cytoplasmic surface of the nuclear membrane, the endoplasmic reticulum, and the outer mitochondrial membrane.
Studies suggest that Bcl-2 family proteins consist of hydrophobic α-helices surrounded by amphipathic α-helices. Several members of the family have transmembrane structural domains at their C-terminus that have a three-dimensional structure consisting of two predominantly hydrophobic central α-helices. They are surrounded by six or seven amphiphilic alpha-helices of different lengths. There are loops between the first two Alpha helices. The Bcl-2 protein structure is strikingly similar to the overall folded structure of bacterial toxins' pore-forming structural domains.
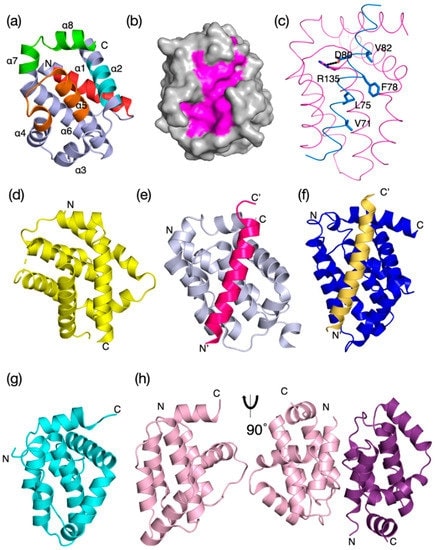 Figure 1. Evolutionary structure conservation in the Bcl-2 family and their complexes. Ribbon representation of the 3D structures of
prosurvival and proapoptotic Bcl-2 family members and their complexes are shown. (Banjara S, et al., 2020)
Figure 1. Evolutionary structure conservation in the Bcl-2 family and their complexes. Ribbon representation of the 3D structures of
prosurvival and proapoptotic Bcl-2 family members and their complexes are shown. (Banjara S, et al., 2020)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| BAK core domain BH3-groove in complex with E. coli lipid: Homo sapiens | Homo sapiens | X-ray diffraction | 2.49 Å | 6UXM |
| BAK core domain BH3-groove-dimer in complex with phosphatidylserine | Homo sapiens | X-ray diffraction | 2.49 Å | 6UXN |
| BAK core domain BH3-groove-dimer in complex with DDM | Homo sapiens | X-ray diffraction | 1.80 Å | 6UXO |
| BAK core domain BH3-groove-dimer in complex with phosphatidylglycerol | Homo sapiens | X-ray diffraction | 2.49 Å | 6UXP |
| BAK core domain BH3-groove-dimer in complex with POPC and C8E4 | Homo sapiens | X-ray diffraction | 1.70 Å | 6UXQ |
| BAK core domain BH3-groove-dimer in complex with LysoPC | Homo sapiens | X-ray diffraction | 1.80 Å | 6UXR |
| Bax core dimer in bicelles (expressed in E. coli) | Homo sapiens | Solution NMR | / | 6L8V |
| Bak transmembrane helix in DPC micelles (expressed in E. coli) | Homo sapiens | Solution NMR | / | 7OFM |
Table 1. Structural research of Pro-apoptotic BCL-2 family.
To understand the molecular mechanisms of apoptosis regulation, it is critical to study the BCL-2 family protein structure. The results of these structural studies will shed light on how BCL-2 family proteins interact with other proteins and their specific functions in the apoptotic signaling pathway. In the future, scientists will be better able to develop effective anti-cancer therapeutic strategies and drug designs by better understanding the structures of these proteins.
Creative Biostructure provides protein structure research services. We are a research service company specializing in protein structural biology research. We provide a range of services related to the structural study of pro-apoptotic BCL-2 family proteins and other proteins. These services include X-ray crystallography, cryo-electron microscopy (Cryo-EM), NMR spectral analysis, and other services. Through the use of advanced experimental and computational techniques, we are committed to helping scientists gain a deeper understanding of the structure and function of pro-apoptotic BCL-2 family proteins. This will facilitate research advances in apoptosis regulation. If you are interested in our services, please contact us for a more detailed description of our services.
References
- Petros AM, et al. Structural biology of the Bcl-2 family of proteins. Biochim Biophys Acta. 2004; 1644(2-3): 83-94.
- Banjara S, et al. The Bcl-2 Family: Ancient Origins, Conserved Structures, and Divergent Mechanisms. Biomolecules. 2020; 10 (1): null.