Structural Research of S-acyltransferases
S-acyltransferase-catalyzed S-palmitoylation of dynamic proteins is one of the fundamental post-translational modifications involved in a wide range of cellular processes and is implicated in human disease-related issues, immune response regulation, and tumor suppression. In recent years, human S-acyltransferases have been recognized as promising drug targets for cancer therapy.
Progress in structural research of DHHC protein S-acyltransferases
The discovery of the DHHC family of S-acyltransferases is one of the breakthroughs in the field of S-acylation. The DHHC proteins contain a conserved protein structural domain of 51 amino acids, which comprises the Asp-His-His-Cys motif embedded in a cysteine-rich structural domain (CRD). Yeast Erf2 and Akr1 represent the core membrane topology. The structures indicate that the DHHC proteins have at least four transmembrane domains (TMDs) with N-terminal and C-terminal exposures to the cytoplasm. Akr1 and its parallel Akr2 homolog expand at the N-terminal end into multiple anchor protein repeats and two additional TMDs. The structures also include a 16-amino acid acyltransferase-conserved c-terminus (PaCCT).
Analysis of the S-acyltransferase mechanism of action
Research on yeast Erf2/Erf4 and mammalian DHHC2/DHHC3 has shown that DHHC proteins have a two-step catalytic mechanism. First, the enzyme undergoes self-acylation using palmitoyl coenzyme a as a donor to form an acylase intermediate. Second, palmitate is transferred to the substrate protein. Mutations in the cysteine in the DHHC motif block both auto-acylation and transfer activity, and mutations in the first histidine block the second step of palmitate transfer. The single transformation supports the hypothesis that DHHC autonylation represents a transient acyl-enzyme transfer intermediate of the two-step ping-pong mechanism. Mass spectrometry analysis of purified DHHC3 confirmed that the DHHC cysteine is modified by palmitate and is the site of the acyl-enzyme intermediate.
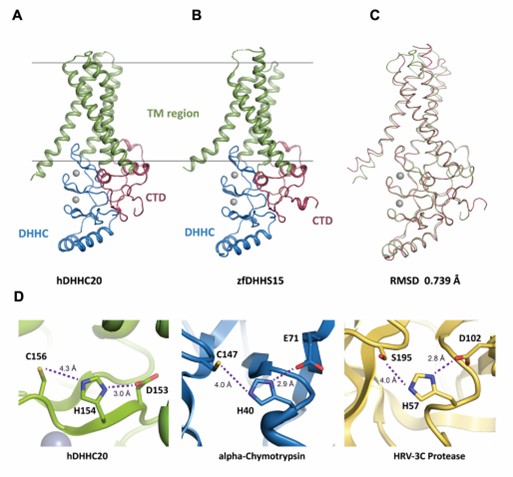 Figure 1. Structure of DHHC-PATs. (Stix R, et al., 2020)
Figure 1. Structure of DHHC-PATs. (Stix R, et al., 2020)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Palmitoyltransferase | Danio rerio | X-ray diffraction | 2.441 Å | 6BMS |
| DHHC20 palmitoyltransferase, space group P63 | Homo sapiens | X-ray diffraction | 2.25 Å | 6BMN |
| DHHC20 palmitoyltransferase, irreversibly inhibited by 2-bromopalmitate | Homo sapiens | X-ray diffraction | 2.95 Å | 6BML |
| Monomeric atSPT-ORM1 (LCB2a-deltaN5) complex | Arabidopsis thaliana | Cryo-EM single particle analysis | 2.8 Å | 7YJO |
| Dimeric atSPT-ORM1 complex | Arabidopsis thaliana | Cryo-EM single particle analysis | 3.2 Å | 7YJK |
| Monomeric atSPT-ORM1 complex | Arabidopsis thaliana | Cryo-EM single particle analysis | 3.2 Å | 7YJM |
| Monomeric atSPT-ORM1 (ORM1-N17A) complex | Arabidopsis thaliana | Cryo-EM single particle analysis | 3.4 Å | 7YJN |
| Serine palmitoyltransferase complex SPTLC1/SPLTC2/ssSPTa | Homo sapiens | Cryo-EM single particle analysis | 3.3 Å | 7K0I |
| Serine palmitoyltransferase complex SPTLC1/SPLTC2/ssSPTa protomer | Homo sapiens | Cryo-EM single particle analysis | 3.1 Å | 7K0J |
| Serine palmitoyltransferase complex SPTLC1/SPLTC2/ssSPTa, 3KS-bound | Homo sapiens | Cryo-EM single particle analysis | 2.6 Å | 7K0K |
| Serine palmitoyltransferase complex SPTLC1/SPLTC2/ssSPTa, myriocin-bound | Homo sapiens | Cryo-EM single particle analysis | 3.4 Å | 7K0L |
| Serine palmitoyltransferase complex SPTLC1/SPLTC2/ssSPTa/ORMDL3, class 1 | Homo sapiens | Cryo-EM single particle analysis | 2.9 Å | 7K0M |
| Serine palmitoyltransferase complex SPTLC1/SPLTC2/ssSPTa/ORMDL3, class 2 | Homo sapiens | Cryo-EM single particle analysis | 3.1 Å | 7K0N |
| Serine palmitoyltransferase complex SPTLC1/SPLTC2/ssSPTa/ORMDL3, class 3 | Homo sapiens | Cryo-EM single particle analysis | 3.1 Å | 7K0O |
| Serine palmitoyltransferase complex SPTLC1/SPLTC2/ssSPTa/ORMDL3, class 4 | Homo sapiens | Cryo-EM single particle analysis | 3.1 Å | 7K0P |
| Serine palmitoyltransferase complex SPTLC1/SPLTC2/ssSPTa/ORMDL3, myriocin-bound | Homo sapiens | Cryo-EM single particle analysis | 3.3 Å | 7K0Q |
| Dimeric SPT-ORMDL3 complex | Homo sapiens | Cryo-EM single particle analysis | 3.8 Å | 6M4N |
| Monomeric SPT-ORMDL3 complex | Homo sapiens | Cryo-EM single particle analysis | 3.4 Å | 6M4O |
| Substrate-bound SPT-ORMDL3 complex | Homo sapiens | Cryo-EM single particle analysis | 3.2 Å | 7CQI |
| Substrate-bound SPT-ORMDL3 complex | Homo sapiens | Cryo-EM single particle analysis | 3.3 Å | 7CQK |
| C6-ceramide-bound SPT-ORMDL3 complex | Homo sapiens | Cryo-EM single particle analysis | 2.9 Å | 7YIU |
| SPT-ORMDL3 (ORMDL3-deltaN2) complex | Homo sapiens | Cryo-EM single particle analysis | 3.1 Å | 7YJ1 |
| SPT-ORMDL3 (ORMDL3-N13A) complex | Homo sapiens | Cryo-EM single particle analysis | 2.9 Å | 7YJ2 |
Table 1. Structural research of S-acyltransferases.
Creative Biostructure, as a leader in structural analysis, understands the essential importance of obtaining high-quality, reliable structural information for protein function research. Our experienced scientists, state-of-the-art cutting-edge equipment, and proven track record of success make us the ideal partner to satisfy your structural biology requirements.
Our X-ray crystallography and cryo-electron microscopy (cryo-EM) technologies enable in-depth structural and mechanism of action investigations of S-acyltransferases to advance the understanding of their structure and function. and action mechanisms to advance the development of S-acyltransferases in cancer therapy. If you are interested in learning more about our services and how we can help you achieve your goals, please feel free to contact us.
References
- Stix R, et al. Structure and Mechanism of DHHC Protein Acyltransferases. J Mol Biol. 2020. 432(18): 4983-4998.
- Gottlieb CD, Linder ME. Structure and function of DHHC protein S-acyltransferases. Biochem Soc Trans. 2017. 45(4): 923-928.
- Zmuda F, Chamberlain LH. Regulatory effects of post-translational modifications on zDHHC S-acyltransferases. J Biol Chem. 2020. 295(43): 14640-14652.