Structural Research of Intramembrane Proteases
Intramembrane proteases are a fascinating class of enzymes that play crucial roles in cellular signaling and protein regulation. The membrane-embedded γ-secretase is one such intramembrane protease responsible for cleaving the transmembrane domains of various signaling proteins, including amyloid precursor protein (APP) and Notch. The malfunction of γ-secretase is closely associated with the pathogenesis of Alzheimer's disease (AD) due to the accumulation of β-amyloid peptides.
A recent study elucidates the cryo-EM structures of human γ-secretase bound to various γ-secretase inhibitors (GSIs) and modulators (GSMs). For instance, the cryo-EM structures revealed that each GSI, such as Semagacestat and Avagacestat, occupies the same general location on presenilin 1 (PS1), which interferes with substrate recruitment. This detailed information on the binding interactions between GSIs and γ-secretase provides crucial insights into the design of substrate-selective inhibitors with improved specificity and efficacy. This study on the GSI L685,458 demonstrated how it directly coordinates with the two catalytic aspartate residues of PS1, thus inhibiting the proteolytic activity of the enzyme. Understanding these mechanisms is crucial for the development of potential therapeutics for AD and other diseases involving γ-secretase dysregulation.
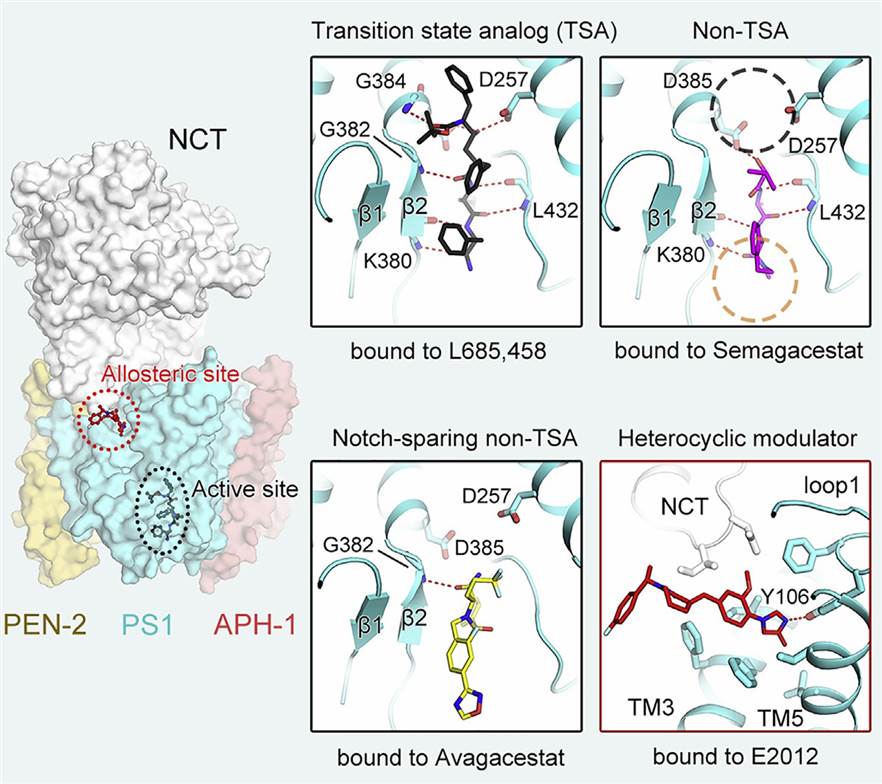 Figure 1. Structural basis of γ-secretase regulation by small molecule drugs. (Yang G, et al., 2021)
Figure 1. Structural basis of γ-secretase regulation by small molecule drugs. (Yang G, et al., 2021)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| GlpG rhomboid-family intramembrane protease (expressed in Escherichia coli) | Escherichia coli | X-ray diffraction | 2.10 Å | 2IC8 |
| GlpG rhomboid-family intramembrane protease (expressed in Escherichia coli) | Escherichia coli | X-ray diffraction | 1.90 Å | 3B45 |
| GlpG rhomboid-family intramembrane protease (expressed in Escherichia coli) | Escherichia coli | X-ray diffraction | 2.50 Å | 2O7L |
| GlpG rhomboid-family intramembrane protease (expressed in Escherichia coli) | Escherichia coli | X-ray diffraction | 2.60 Å | 2NRF |
| GlpG rhomboid-family intramembrane protease (expressed in Escherichia coli) | Escherichia coli | X-ray diffraction | 2.30 Å | 2IRV |
| GlpG rhomboid-family intramembrane protease (expressed in Escherichia coli) | Escherichia coli | X-ray diffraction | 1.65 Å | 2XOV |
| GlpG rhomboid-family intramembrane protease with lipids (expressed in Escherichia coli) | Escherichia coli | X-ray diffraction | 1.70 Å | 2XTV |
| GlpG rhomboid-family intramembrane protease with a mechanism-based inhibitor (expressed in Escherichia coli) | Escherichia coli | X-ray diffraction | 2.30 Å | 3TXT |
| GlpG rhomboid-family intramembrane protease in complex with phosphonofluoridate inhibitor (expressed in Escherichia coli) | Escherichia coli | X-ray diffraction | 2.60 Å | 3UBB |
| GlpG N-terminal cytoplasmic domain (expressed in Escherichia coli) | Pseudomonas aeruginosa PAO1 | Solution NMR | / | 2GQC |
| GlpG N-terminal cytoplasmic domain (expressed in Escherichia coli) | Escherichia coli | Solution NMR | / | 2LEP |
| GlpG N-terminal cytoplasmic domain (expressed in Escherichia coli) | Escherichia coli | X-ray diffraction | 1.35 Å | 4HDD |
| GlpG rhomboid-family intramembrane protease in complex with inhibitor Ac-IATA-cmk (expressed in Escherichia coli) | Escherichia coli | X-ray diffraction | 2.10 Å | 4QO2 |
| GlpG rhomboid-family intramembrane protease in complex with Ac-VRMA-CHO (expressed in Escherichia coli) | Escherichia coli | X-ray diffraction | 2.30 Å | 5F5B |
| GlpG rhomboid intramembrane protease snapshots, I1 (expressed in Escherichia coli) | Escherichia coli | X-ray diffraction | 2.30 Å | 6PJ4 |
| GlpG rhomboid-family intramembrane protease (expressed in Escherichia coli) | Haemophilus influenzae 86-028NP | X-ray diffraction | 2.20 Å | 2NR9 |
| GlpG rhomboid-family intramembrane protease (expressed in Escherichia coli) | Haemophilus influenzae | X-ray diffraction | 2.84 Å | 3ODJ |
| Site-2 Protease (S2P). Intramembrane Metalloprotease (expressed in Escherichia coli) | Methanocaldococcus jannaschii | X-ray diffraction | 3.30 Å | 3B4R |
| Signal Peptide Peptidase (SppA), native protein (expressed in Escherichia coli) | Escherichia coli | X-ray diffraction | 2.55 Å | 3BF0 |
| Signal Peptide Peptidase (SppA) (expressed in Escherichia coli) | Bacillus subtilis | X-ray diffraction | 2.37 Å | 3RST |
| Signal Peptide Peptidase (SppA) K199A mutant showing C-terminal peptide bound in eight active sites (expressed in Escherichia coli) | Bacillus subtilis subsp. subtilis str. 168 | X-ray diffraction | 2.39 Å | 4KWB |
| Lipoprotein signal peptidase II (expressed in Escherichia coli) | Pseudomonas aeruginosa PAO1 | X-ray diffraction | 2.80 Å | 5DIR |
| LspA lipoprotein signal peptide peptidase II in complex with globomycin (expressed in Escherichia coli) | Staphylococcus aureus | X-ray diffraction | 1.92 Å | 6RYO |
| PSH presenilin/SPP homologue aspartate protease (C222 space group) (expressed in Escherichia coli) | Methanoculleus marisnigri JR1 | X-ray diffraction | 3.32 Å | 4HYG |
| FlaK preflagellin aspartyl protease (expressed in Escherichia coli) | Methanococcus maripaludis | X-ray diffraction | 3.60 Å | 3S0X |
| CAAX protease Ste24p (expressed in Saccharomyces cerevisiae) | Saccharomyces mikatae | X-ray diffraction | 3.10 Å | 4IL3 |
| CAAX protease ZMPSTE24 (expressed in Spodoptera frugiperda) | Homo sapiens | X-ray diffraction | 3.40 Å | 4AW6 |
| CAAX protease ZMPSTE24 with bound phosphoramidon (expressed in Saccharomyces cerevisiae) | Homo sapiens | X-ray diffraction | 3.85 Å | 6BH8 |
| CAAX protease Rce1 (expressed in Escherichia coli) | Mus musculus, Methanococcus maripaludis | X-ray diffraction | 2.50 Å | 4CAD |
| γ-secretase (expressed in HEK293S cells) | Homo sapiens | Cryo-EM single particle analysis | 4.40 Å | 4UIS |
| γ-secretase (expressed in HEK293F cells) | Homo sapiens | Cryo-EM single particle analysis | 3.40 Å | 5A63 |
| γ-secretase nicastrin-a transmembrane domain in SDS (expressed in Escherichia coli) | Homo sapiens | Solution NMR | / | 2N7Q |
| γ-secretase in complex with Notch-100 (expressed in HEK293 cells) | Homo sapiens | Cryo-EM single particle analysis | 2.70 Å | 6IDF |
| γ-secretase cross-linked complex with APP-C83 (expressed in HEK293 cells) | Homo sapiens | Cryo-EM single particle analysis | 2.60 Å | 6IYC |
| γ-secretase substrate: TREM2 transmembrane helix in DPC micelles (expressed in Escherichia coli) | Homo sapiens | Solution NMR | / | 6Z0G |
| γ-secretase with bound Semagacestat (expressed in HEK293S cells) | Homo sapiens | Cryo-EM single particle analysis | 3.00 Å | 6LR4 |
| γ-secretase nicastrin extracellular domain (expressed in Spodoptera frugiperda) | Dictyostelium purpureum | X-ray diffraction | 1.95 Å | 4R12 |
| Signal Peptidase Complex Paralog A (SPC-A) (expressed in HEK293 cells) | Homo sapiens | Cryo-EM single particle analysis | 4.90 Å | 7P2P |
| Signal peptidase complex subunit 1 (expressed in Komagataella pastoris) | Aequorea victoria, Gallus gallus | X-ray diffraction | 2.43 Å | 6WVE |
| PGAM5 WT transmembrane domain (TMD) | Homo sapiens | Solution NMR | / | 7QAM |
Table 1. Structural research of intramembrane proteases.
As a reputable company specializing in structural analysis services, Creative Biostructure has been instrumental in advancing our understanding of intramembrane proteases. Our team of expert scientists has extensive experience in cryo-EM studies, enabling us to elucidate high-resolution structures of complex membrane proteins.
Utilizing cutting-edge techniques such as X-ray crystallography, nuclear magnetic resonance (NMR) spectroscopy, and cryo-electron microscopy (cryo-EM), our team of skilled researchers excels in elucidating the intricate three-dimensional structures of membrane protein complexes. This expertise empowers us to uncover valuable insights into their functional mechanisms and interactions, making substantial contributions to the understanding of biological processes. Contact us today to unlock the full potential of our capabilities, elevating your research pursuits and propelling you towards the achievement of your scientific ambitions.
References
- Cho S, et al. Ten catalytic snapshots of rhomboid intramembrane proteolysis from gate opening to peptide release. Nature Structural & Molecular Biology. 2019, 26(10): 910-918.
- Olatunji S, et al. Structures of lipoprotein signal peptidase II from Staphylococcus aureus complexed with antibiotics globomycin and myxovirescin. Nature Communications. 2020, 11(1): 140.
- Yang G, et al. Structural basis of γ-secretase inhibition and modulation by small molecule drugs. Cell. 2021, 184(2): 521-533. e14.