Structural Research of Reoviridae
The Reoviridae (now called Sedoreoviridae) are the largest and most diverse family of double-stranded RNA (dsRNA) viruses, comprising 12 identified genera. Reovirus infections occur frequently in humans. Of these, Rotaviruses can cause severe diarrhea and intestinal distress in children. Orthoreoviruses are associated with the celiac disease. In addition, in agricultural environments, reovirus causes plant stunting and necrosis, leading to significant crop losses. Therefore, scientists have gained an in-depth understanding of reovirus molecular structure in recent years, revealing the mechanism of virus entry, replication, assembly, and evasion of host immune response, which lays the foundation for the development of intervention and control strategies.
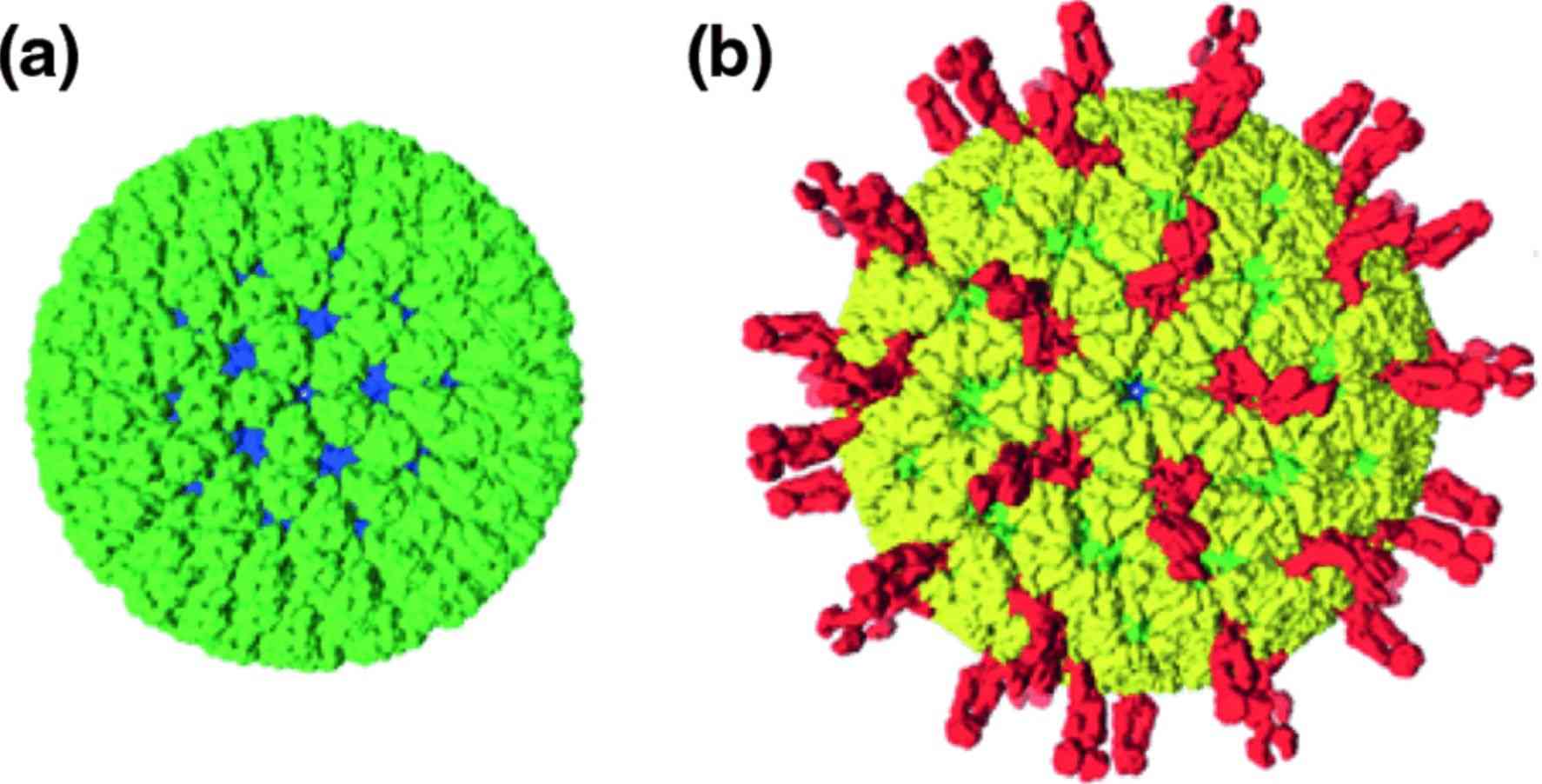 Figure 1. Rotaviruses particle structure. (Matthijnssens J, et al, 2022)
Figure 1. Rotaviruses particle structure. (Matthijnssens J, et al, 2022)
Structural Overview of Reoviruses
The genome of Reoviruses consists of 9-12 segments, which are classified according to their size into L (large) encoding the λ-protein, M (medium) encoding the μ-protein, and S (small) encoding the σ-protein. The positive strand of each bicistronic body is modified with a 5' terminal type 1 cap structure and has no 3'-poly(A) tail. Reoviruses are unenveloped and have an icosahedral capsid consisting of an outer (T = 13) and an inner (T = 2) protein shell. The inner capsid is morphologically similar and consists of 120 thin crescent-shaped protein copies that exhibit distinct modifications. Reoviruses have a unique structure that contains glycosylated spiny proteins on their surface.
Structural Features of Reoviridae Family Members
Using cryo-electron microscopy (cryo-EM), researchers have determined the structure of the mud crab reovirus (MCRV), a 12-segmented dsRNA virus, a member of the Reoviridae family, with icosahedral symmetry (3.1 Å) and without imposing icosahedral symmetry (3.4 Å). The structure reveals a two-layered organization of the major coat protein, with the outer T = 13 composed of VP12 trimers and a unique VP11 clamp, and the inner T = 1 composed of VP3 dimers. In addition, 10 RNA-dependent RNA polymerases (RdRp) are below the VP3 layer, but of the 5-fold axis and arranged with D5 symmetry, unlike other members of the Reoviridae family. The N-terminus of VP3 has four conformations, two anchoring RdRps, and two others associated with genome organization and capsid stability.
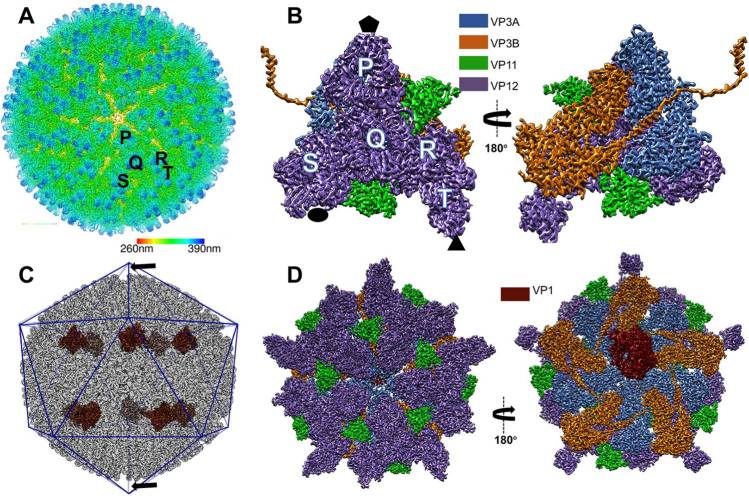 Figure 2. The overall structure of mud crab reovirus (MCRV). (Zhang Q, et al, 2023)
Figure 2. The overall structure of mud crab reovirus (MCRV). (Zhang Q, et al, 2023)
Creative Biostructure offers a range of high-quality virus-like particles (VLPs) products and associated structure elucidation services that are of great importance in virology, vaccine development, and therapeutic research. Our VLPs mimic the structural features of actual viruses but lack the genetic material, making them an invaluable tool for researching the structural and functional properties of viruses.
| Cat No. | Product Name | Virus Name | Source | Composition |
| CBS-V542 | African horse sickness virus VLP (VP5; VP2; VP7; VP3 Proteins) | African horse sickness virus | Plant recombinant | VP5; VP2; VP7; VP3 |
| CBS-V653 | Human rotavirus VLP (VP2; VP4; VP6; VP7 Proteins) | Rotavirus | Insect cell recombinant | VP2; VP4; VP6; VP7 |
| CBS-V737 | Rotavirus VLP (VP8 Protein) | Rotavirus | E. coli recombinant | VP8 |
| Explore All Reoviridae Virus-like Particle Products | ||||
Using cutting-edge techniques such as cryo-electron microscopy (cryo-EM) and X-ray crystallography to capture high-resolution structural data, Creative Biostructure can determine the three-dimensional structure of viruses, providing essential insights into molecular structure, viral proteins, and interactions with the host.
We also prioritize personalized support and collaboration with our clients. We understand the unique requirements of each research project and work closely with clients to tailor solutions to specific needs. Feel free to contact us for more details.
References
- Matthijnssens J, et al. ICTV Virus Taxonomy Profile: Sedoreoviridae 2022. J Gen Virol. 2022. 103(10): 10.1099/jgv.0.001782.
- Zhang Q, et al. The structure of a 12-segmented dsRNA reovirus: New insights into capsid stabilization and organization. PLoS Pathog. 2023. 19(4): e1011341.