Structural Research of Membrane-Shaping Proteins
Membrane-shaping proteins are a class of proteins that regulate the morphology and dynamics of biofilms. They are involved in some cellular processes, including endocytosis, exocytosis, cell division, and organelle biogenesis.
Cryo-electron microscopy and X-ray diffraction are usually used to study the structure of membrane Shaping proteins. Both of these technologies can provide high-resolution images of protein structure, which is helpful in determining the exact shape of protein complexes and understand their functions.
A study published in 2022 revealed the structural basis of how membrane proteins such as caveolin interact with cell membranes using low-temperature electron microscopy. The human Caveolin-1 complex comprises 11 plastids arranged tightly on a flat membrane surface. This study reveals how the critical regions of caveolin-1 contribute to its membrane remodeling function in response to physiological and environmental cues.
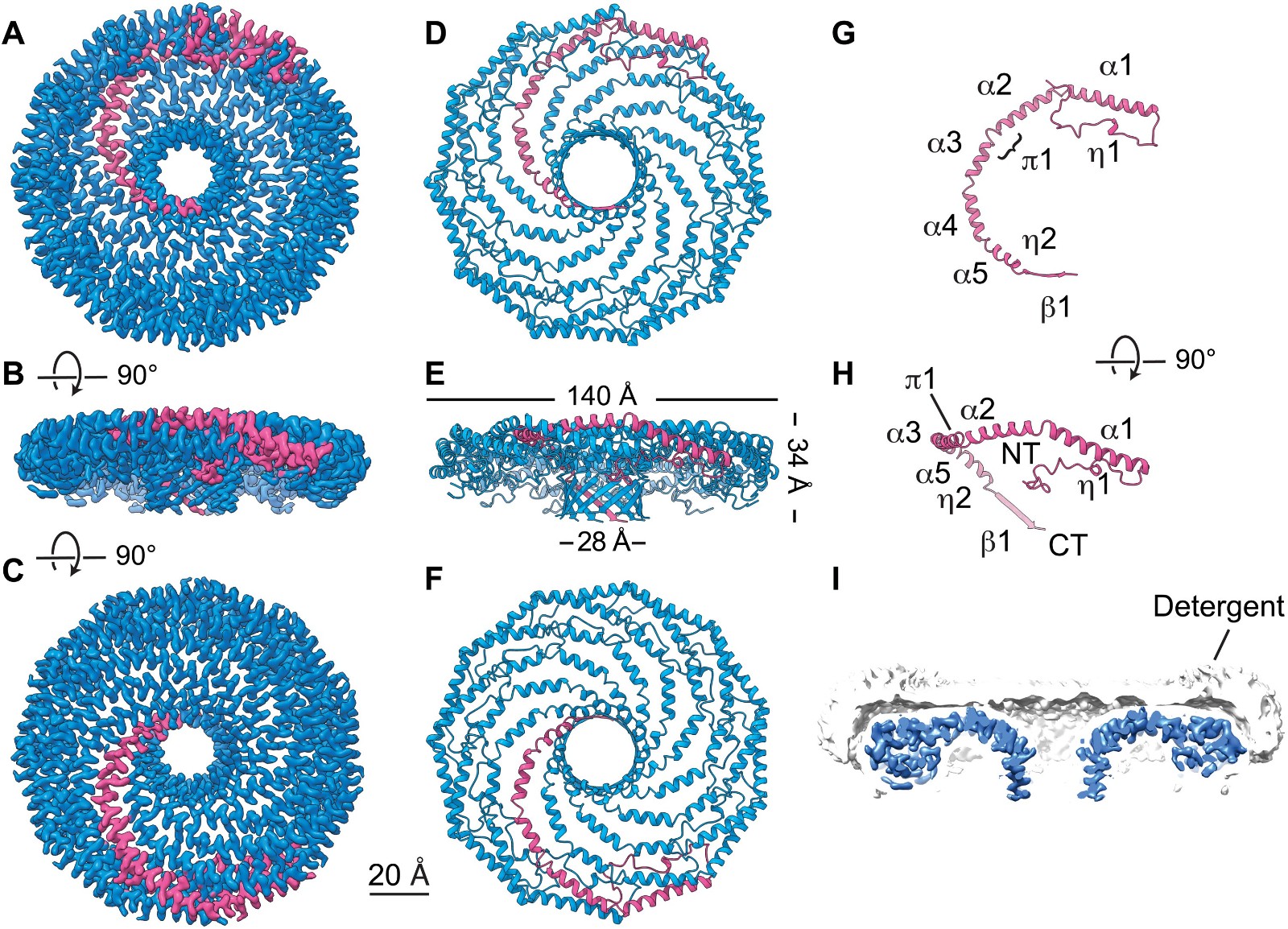 Figure 1. Cryo-EM structure of the 8S Cav1 complex. (PORTA , et al., 2022)
Figure 1. Cryo-EM structure of the 8S Cav1 complex. (PORTA , et al., 2022)
To study the structure and assembly of GTPase Mgm1 for mitochondrial membrane remodeling, in an article published in 2019, researchers showed the crystal and electronic low-temperature tomography structure of Mgm1 from Chaetomium thermophilum, revealing the molecular basis of the assembly of Mgm1 into filaments, and provided a model of how the rearrangement of these filaments can induce inner mitochondrial membrane remodeling.
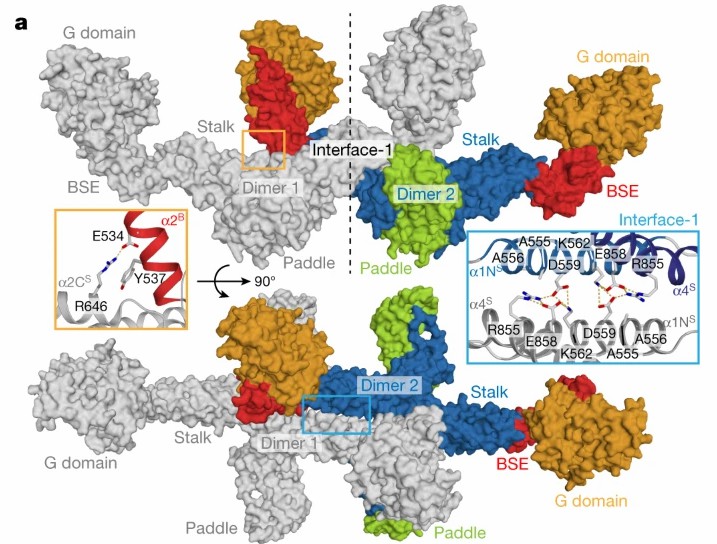 Figure 2. Assembly mechanism of Mgm1. (FAELBER, et al., 2019)
Figure 2. Assembly mechanism of Mgm1. (FAELBER, et al., 2019)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| IST1-CHMP1B ESCRT-III copolymer | Homo sapiens | Cryo-EM | 4.00 Å | 3JC1 |
| ESCRT-III filament composed of CHMP1B, membrane-bound | Homo sapiens | Cryo-EM | 6.20 Å | 6TZ9 |
| ESCRT-III filament composed of CHMP1B+IST1 (right-handed) | Homo sapiens | Cryo-EM | 3.20 Å | 6TZ4 |
| ESCRT-III filament composed of CHMP1B+IST1 (left-handed) | Homo sapiens | Cryo-EM | 3.10 Å | 6TZ5 |
| filament composed of IST1 NTD, R16E,K27E mutant | Homo sapiens | Cryo-EM | 7.20 Å | 6TZA |
| CHMP2A-CHMP3 ESCRT-III copolymer, 430 Å diameter | Homo sapiens | Cryo-EM | 3.30 Å | 7ZCG |
| CHMP2A-CHMP3 ESCRT-III copolymer, 410 Å diameter | Homo sapiens | Cryo-EM | 3.60 Å | 7ZCH |
| ESCRT-III Snf7 core domain, conformation A | Saccharomyces cerevisiae S288C | X-ray diffraction | 2.40 Å | 5FD7 |
| ESCRT-III Snf7 core domain, conformation B | Saccharomyces cerevisiae S288C | X-ray diffraction | 1.60 Å | 5FD9 |
| Shrub, fly ortholog of ESCRT-III SNF7/CHMP4B | Drosophila melanogaster | X-ray diffraction | 2.76 Å | 5J45 |
| Retromer Vps26-Vps29-Vps35 complex | Thermochaetoides thermophila DSM 1495 | Cryo-EM tomography | 11.4 Å | 6H7W |
| Vps29 | Thermochaetoides thermophila | X-ray diffraction | 1.52 Å | 5W8M |
| Seipin lipid droplet-formation protein | Drosophila melanogaster | Cryo-EM | 4.00 Å | 6MLU |
| Seipin lipid droplet-formation protein | Saccharomyces cerevisiae | Cryo-EM | 3.45 Å | 7RSL |
| Mgm1 mitochondrial remodeling GTPase | Thermochaetoides thermophila DSM 1495 | X-ray diffraction | 3.6 Å | 6QL4 |
| s-Mgm1 decorating the outer surface of tubulated lipid membranes | Thermochaetoides thermophila | Cryo-EM | 14.7 Å | 6RZT |
| s-Mgm1 decorating the outer surface of tubulated lipid membranes in the GTPγS bound state | Thermochaetoides thermophila DSM 1495 | Cryo-EM | 14.7 Å | 6RZU |
| s-Mgm1 decorating the inner surface of tubulated lipid membranes | Thermochaetoides thermophila DSM 1495 | Cryo-EM | 20.6 Å | 6RZV |
| s-Mgm1 decorating the inner surface of tubulated lipid membranes in the GTPγS bound state | Thermochaetoides thermophila DSM 1495 | Cryo-EM | 18.8 Å | 6RZW |
| Sorting nexin (SNX)-BAR Mvp1 tetramer | Saccharomyces cerevisiae W303 | Cryo-EM | 4.20 Å | 6Q0X |
| myelin P2 protein, N2D variant | Homo sapiens | X-ray diffraction | 1.65 Å | 6XU5 |
| P2 protein K3N mutant | Homo sapiens | X-ray diffraction | 2.70 Å | 6XU9 |
| Endophilin B1 N-BAR helical scaffold protein | Mus musculus | Cryo-EM | 9.00 Å | 6UP6 |
| Sorting nexin (SNX) protein SNX1, helical array | Mus musculus | Cryo-EM | 9.00 Å | 7D6D |
| Coat protein complex II (COPII) on membranes, outer coat right-handed rod, class1 | Saccharomyces cerevisiae S288C | Cryo-EM | 40.0 Å | 6ZG5 |
| Metazoan membrane-assembled retromer | Homo sapiens | Cryo-EM | 8.90 Å | 7BLN |
| Atlastin-1 GTPase | Homo sapiens | X-ray diffraction | 2.20 Å | 6XJN |
| Caveolin-1 complex | Homo sapiens | Cryo-EM | 3.40 Å | 7SC0 |
| EHD4 complex | Mus musculus | Cryo-EM tomography | 7.60 Å | 7SOX |
Table 1. Structural research of membrane-shaping proteins.
Creative Biostructure is committed to providing exceptional X-ray crystallography and cryo-electron microscopy(cryo-EM) services, which are effective methods for investigating the three-dimensional structures of membrane proteins. We employ advanced detection equipment and methodologies to deliver optimal client services and outcomes. Our laboratory staff are extensively trained and certified and possess the expertise and proficiency to ensure precise and dependable data delivery.
References
- FAELBER K, et al. Structure and assembly of the mitochondrial membrane remodeling GTPase MGM1. Nature, 2019, 571(7765): 429–433.
- ARLT H, et al. Seipin forms a flexible cage at lipid droplet formation sites. Nature Structural & Molecular Biology, 2022, 29(3): 194–202.
- SUI X, et al. Cryo–electron microscopy structure of the lipid droplet–formation protein seipin. Journal of Cell Biology, 2018, 217(12): 4080–4091.
- KOVTUN O, et al. Structure of the membrane-assembled retromer coat determined by cryo-electron tomography. Nature, 2018, 561(7724): 561–564.
- MCMILLAN B J, et al. Electrostatic interactions between elongated monomers drive filamentation of drosophila shrub, a metazoan ESCRT-III protein. Cell Reports, 2016, 16(5): 1211–1217.
- TANG S, et al. Structural basis for activation, assembly and membrane binding of ESCRT-III Snf7 filaments. eLife, 2015, 4.
- AZAD K, et al. Structural basis of CHMP2A–CHMP3 ESCRT-III polymer assembly and membrane cleavage. Nature Structural & Molecular Biology, 2023, 30(1): 81–90.
- NGUYEN H C, et al. Membrane constriction and thinning by sequential ESCRT-III Polymerization. Nature Structural & Molecular Biology, 2020, 27(4): 392–399.
- MCCULLOUGH J, et al. Structure and membrane remodeling activity of ESCRT-III helical polymers. Science, 2015, 350(6267): 1548–1551.
- MELO A A, et al. Cryo-electron tomography reveals structural insights into the membrane remodeling mode of dynamin-like EHD filaments. Nature Communications, 2022, 13(1).
- PORTA J C, et al. Molecular architecture of the human caveolin-1 complex. Science Advances, 2022, 8(19).
- KELLY C M, et al. The hypervariable region of atlastin-1 is a site for intrinsic and extrinsic regulation. Journal of Cell Biology, 2021, 220(11).
- LENEVA N, et al. Architecture and mechanism of metazoan retromer:SNX3 tubular coat assembly. Science Advances, 2021, 7(13).
- HUTCHINGS J, et al. Structure of the complete, membrane-assembled copii coat reveals a complex interaction network. Nature Communications, 2021, 12(1).
- ZHANG Y, et al. Structural insights into membrane remodeling by SNX1. Proceedings of the National Academy of Sciences, 2021, 118(10).
- BHATT V S, et al. Amphipathic motifs regulate N-bar protein endophilin B1 auto-inhibition and drive membrane remodeling. Structure, 2021, 29(1).
- RUSKAMO S, et al. Cryo-Em, X-ray diffraction, and atomistic simulations reveal determinants for the formation of a supramolecular myelin-like proteolipid lattice. Journal of Biological Chemistry, 2020, 295(26): 8692–8705.
- SUN D, et al. The cryo-EM structure of the SNX–bar MVP1 tetramer. Nature Communications, 2020, 11(1).