All-atom Mempro™ Nanodisc Dynamics Simulation
With the development of the magic Mempro™ Membrane Protein platform in the recent years, Creative Biostructure has established a novel all-atom molecular dynamics simulation platform for our advanced Mempro™ Nanodisc technology to better understand the structural and functional aspects of membrane proteins, lipid-protein interactions, and protein-protein interaction in a native-like lipid environment. All-atom Mempro™ Nanodisc Dynamics Simulation, which belongs to the Mempro™ Nanodisc Computational Studies, has developed a convincing method to mimic the real lipids environment for membrane proteins. The introduction of AT-MD approach makes Mempro™ Nanodisc platform more valuable and with bright prospects in a variety of applications. All-atom simulation has been applied in various research fields such as membrane models, chemical compounds, DNAs, and proteins, and this method has already made a series of achievements because of the high resolution and the accurate prediction. Nowadays, the development trend also shows that the computer industry has great potentials in the membrane proteins research with its native lipid bilayers around.
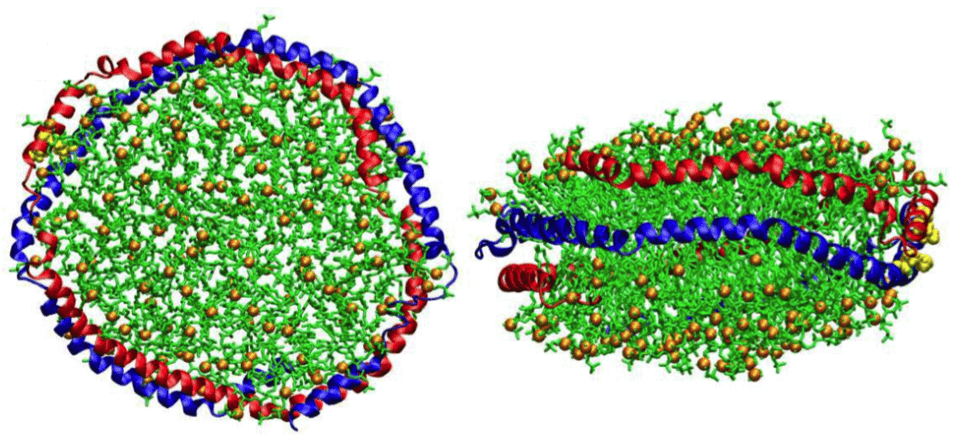 Figure 1. Schematic diagram of the nanodisc in all-atom simulation (left, top view and right, side view). Red and blue represent the two monomers 1 and 2 of aMSP1Δ, respectively. DMPC and phosphate headgroups are colored in green and orange, and lysine residues at position 90 are highlighted in yellow. Water molecules are not shown in the figure. (J. Phys. Chem. B 2015)
Figure 1. Schematic diagram of the nanodisc in all-atom simulation (left, top view and right, side view). Red and blue represent the two monomers 1 and 2 of aMSP1Δ, respectively. DMPC and phosphate headgroups are colored in green and orange, and lysine residues at position 90 are highlighted in yellow. Water molecules are not shown in the figure. (J. Phys. Chem. B 2015)
Mempro™ Nanodisc technology is an advanced tool for functional and structural studies of membrane proteins, while many experimental studies of nanodiscs is limited to investigate the properties of membrane scaffold proteins and membrane proteins, the structural details of nanodisc self-assembly are still missing. In order to solve these problems, Creative Biostructure has established the All-atom Mempro™ Nanodisc Dynamics Simulation platform, which can provide much more details of the structure and interactions of the MSPs with the lipids and with themselves. Compared with traditional experimental methods, All-atom Mempro™ Nanodisc Dynamics Simulation has a variety advantages as follows: 1) the processes and results are not affected by environmental factors since this approach can mimic a real membrane environment; 2) the resolution observed is higher and easily to be observed; 3) all the calculation process is done by the computer which ensures the precise and accurate data.
In All-atom Mempro™ Nanodisc Dynamics Simulation, the systems were performed by Gromacs software and visualized by VMD software, appropriate temperature and pressure were required depend on the specific system set-up, and the particle-mesh Ewald (PME) method and harmonic position restraints were also used. Creative Biostructure can easily use the backmapping algorithm to convert the coarse-grained conformation into an atomistic model. Various phospholipids can be constructed and applied to build the nanodisc models with desired membrane scaffold proteins.
All-atom Mempro™ Nanodisc Dynamics Simulation has shown to be a desired alternative to explore membrane proteins and lipid bilayers. Creative Biostructure also provides a broad range of Mempro™ Nanodisc products and services. Please see the related sections for further information and feel free to contact us for a detailed quote.
References:
Debnath, et al. (2015). Structure and Dynamics of Phospholipid Nanodiscs from All-Atom and Coarse-Grained Simulations. Journal of Physical Chemistry B, 119(23): 6991-7002.
I. Siuda, et al. (2015). Molecular Models of Nanodiscs.Journal of Chemical Theory and Computation, 11: 4923−4932.
Rzepiela, A. J., et al. (2010). Reconstruction of atomistic details from coarse-grained structures. J. Comput. Chem. 31, 1333-1343.